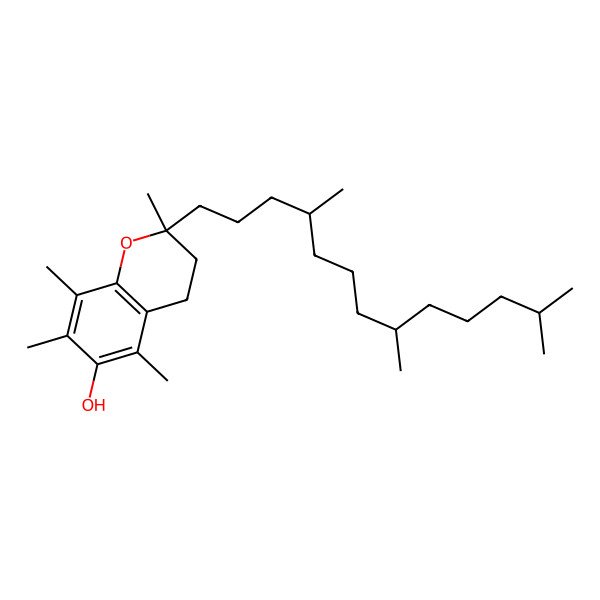
| alpha-Tocopherol |
| D-alpha-Tocopherol |
| 59-02-9 |
| 5,7,8-Trimethyltocol |
| (+)-alpha-Tocopherol |
| alpha Tocopherol |
| Phytogermine |
| TOCOPHEROL |
| 2074-53-5 |
| (R,R,R)-alpha-Tocopherol |
| Syntopherol |
| Viteolin |
| Eprolin |
| Esorb |
| a-Tocopherol |
| (2R,4'R,8'R)-alpha-Tocopherol |
| Tocopherol alpha |
| dl-a-Tocopherol |
| Profecundin |
| Denamone |
| Epsilan |
| Tokopharm |
| Vascuals |
| Viprimol |
| Etavit |
| alpha-Tokoferol |
| Evion |
| alpha-Tocopherol, D- |
| alpha-Vitamin E |
| Vitamin E alpha |
| Eprolin S |
| Viterra E |
| E Prolin |
| D-alpha tocopherol |
| Aquasol E |
| Lan-E |
| Med-E |
| Antisterility vitamin |
| alpha-Tocopherol acid |
| Tenox GT 1 |
| Vi-E |
| Rhenogran Ronotec 50 |
| (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL |
| Covitol F 1000 |
| Waynecomycin |
| E 307 (tocopherol) |
| (R)-2,5,7,8-Tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)chroman-6-ol |
| Vitamin Ea |
| ido-E |
| Tocopherol (R,S) |
| CCRIS 3588 |
| 1406-18-4 |
| CHEBI:18145 |
| Vitamin-E |
| 2,5,7,8-Tetramethyl-2-(4',8',12'-trimethyltridecyl)-6-chromanol |
| HSDB 2556 |
| E-Vimin |
| EINECS 200-412-2 |
| NSC 20812 |
| (all-R)-alpha-Tocopherol |
| Evitaminum |
| Almefrol |
| E307 |
| Emipherol |
| Etamican |
| Vitayonon |
| Ilitia |
| (+-)-Med-E |
| UNII-N9PR3490H9 |
| (2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydrochromen-6-ol |
| H4N855PNZ1 |
| BPBio1_000362 |
| 18920-62-2 |
| Vitaplex E |
| Covi-ox |
| DTXSID0026339 |
| E 307 |
| .ALPHA.-TOCOPHEROL, D- |
| Spavit E |
| EC 200-412-2 |
| alpha-D-Tocopherol |
| Endo E |
| N9PR3490H9 |
| Vita E |
| EINECS 215-798-8 |
| EINECS 218-197-9 |
| a-Vitamin E |
| NSC 82623 |
| (2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydro-2H-chromen-6-ol |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-, (2R)- |
| (+-)-alpha-Tocopherol |
| C29H50O2 |
| (+/-)-alpha-Tocopherol |
| rel-alpha-Vitamin E |
| 3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-benzopyran-6-ol |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-, (2R)-rel- |
| CCRIS 5853 |
| DTXCID706339 |
| .alpha.-Tocopherol |
| d-a-tocopherol |
| EINECS 233-466-0 |
| MFCD00072045 |
| RRR-alpha-tocopherol |
| BRN 0094012 |
| CAS-59-02-9 |
| (2R*(4R*,8R*))-(1)-3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-benzopyran-6-ol |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-, (2R)- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, (2R-(2R*(4R*,8R*)))- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, [2R-[2R*(4R*,8R*)]]- |
| SMR000471844 |
| VIV |
| .Alpha.-tocopherol, DL- |
| 3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-ol |
| DTXSID8021355 |
| Phytogermin |
| Palmvtee |
| alpha-Tocoferol |
| Vitamin Ealpha |
| a- tocopherol |
| NSC-20812 |
| ?-Tocopherols |
| Alpha-tocopherols |
| a-D-Tocopherol |
| Pheryl-E |
| DL--Tocopherol |
| Vita plus E |
| rel--Vitamin E |
| -Vitamin E |
| d-..-Tocopherol |
| Vitamin e d-alpha |
| NCGC00016688-02 |
| (+)--tocopherol |
| Prestwick_653 |
| .alpha.-Vitamin E |
| Envirose 100 |
| (+)-a-Tocopherol |
| COVIREL |
| RRR-alpha-tocopheryl |
| Vitamin E [USP] |
| ()-alpha-Tocopherol |
| delta-alpha-tocopherol |
| alpha-delta-Tocopherol |
| Tocopherol, d-alpha- |
| CONTROX VP |
| EPROLIN-S |
| Vitamin E (D-form) |
| TENOX GT |
| CHEMBL47 |
| (R,R,R)-a-Tocopherol |
| Prestwick3_000404 |
| (+)- alpha -Tocopherol |
| (+)-.alpha.-Tocopherol |
| bmse000600 |
| R,r,r-.alpha.-tocopherol |
| alpha-TOCOPHEROL (II) |
| SCHEMBL3097 |
| COVI-OX T 30P |
| RIKKI N 70 |
| UNII-H4N855PNZ1 |
| BIDD:PXR0174 |
| COVI-OX T 50 |
| BSPBio_000328 |
| E-MIX 40A |
| MLS001066396 |
| MLS001335981 |
| MLS001335982 |
| BIDD:ER0562 |
| INS NO.307A |
| T1539_SIGMA |
| DTXCID201355 |
| INS-307A |
| (+)-ALPHA-TOCOPHEROL- |
| ALPHA-TOCOPHEROL [HSDB] |
| Vitamin e (as alpha tocopherol) |
| (2R,4'R,8'R)-a-Tocopherol |
| .ALPHA.-TOCOPHEROL [MI] |
| ((addition))-(alpha)-Tocopherol |
| (2R,4'R,8'R)-2,5,7,8-Tetramethyl-2-(4',8',12'-trimethyltridecyl)-6-chromanol |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl- 2-(4,8,12-trimethyltridecyl)- |
| 3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1- -benzopyran-6-ol |
| ALPHA TOCOPHEROL (USP-RS) |
| (+)--Tocopherol; D--Tocopherol |
| HMS2096A10 |
| HMS2231G08 |
| C29H50O2 (D-alpha-tocopherol) |
| 3, 4- dihydro- 2, 5, 7, 8- tetramethyl- 2- (4, 8, 12- trimethyltridecyl)- 2H- benzopyran- 6- ol |
| D-ALPHA TOCOPHEROL [MART.] |
| HY-N0683 |
| Tox21_110563 |
| Tox21_113208 |
| Tox21_202081 |
| BDBM50458513 |
| E-307A |
| LMPR02020001 |
| AKOS004910417 |
| CS-8161 |
| CS-O-02555 |
| DB00163 |
| LS-7727 |
| (2R)-2-((4R,8R)-4,8,12-trimethyltridecyl)-2,5,7,8-tetramethylchroman-6-ol |
| NCGC00142625-01 |
| NCGC00142625-04 |
| NCGC00142625-05 |
| NCGC00142625-06 |
| NCGC00142625-07 |
| NCGC00142625-10 |
| NCGC00259630-01 |
| ALPHA-TOCOPHEROL, UNSPECIFIED FORM |
| AS-13990 |
| J24.260H |
| rel-(+)--Tocopherol; rel-D--Tocopherol |
| J203.513H |
| LS-173092 |
| E-307 |
| T2309 |
| C02477 |
| F82497 |
| d-alpha, d-beta, d-gamma & d-delta tocopherols |
| EN300-7417123 |
| Q158348 |
| Q-201932 |
| W-107596 |
| W-109164 |
| Z2235811339 |
| 07AA93F0-3339-4EEC-B50B-ADB70F657087 |
| 3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1--benzopyran-6-ol |
| (2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydro-2H-1-benzopyran-6-ol |
| (2R)-3,4-Dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-2H-1-benzopy-ran-6-ol |
| (2R)-3,4-Dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-2H-1-benzopyran-6-ol |
| (R)-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-3,4-dihydro-2H-chromen-6-ol |
| [2R*(4R*,8R*)]-(+-)-3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-benzopyran-6-ol |
| 2H-1-benzopirano-6-ol, 3,4-dihidro-2,5,7,8-tetrametil-2-[(4R, 8R)-4,8,12-trimetiltridecil]-, (2R)- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl- 2-(4,8,12-trimethyltridecyl)-, [2R-[2R(4R,8R)]]- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, 2R- 2R*(4R*,8R*) - |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, (2R*(4R*,8R*))-(+-)- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, [2R*(4R*,8R*)]-(.+-.)- |
| 2H-1-Benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, [2R*(4R*,8R*)]-(.+.)- |
| rel-(2R)-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-3,4-dihydro-2H-chromen-6-ol |
|
There are more than 10 synonyms. If you wish to see them all click here.
|