| 144-68-3 |
| Anchovyxanthin |
| all-trans-Zeaxanthin |
| Zeaxanthol |
| Xanthophyll 3 |
| Beta,beta-carotene-3,3'-diol |
| Luteinofta |
| Optisharp |
| Zeagold |
| 3R,3'R-Zeaxanthin |
| trans-Zeaxanthin |
| (3R,3'R)-Zeaxanthin |
| beta-carotene-3,3'-diol |
| (3R,3'R)-beta,beta-Carotene-3,3'-diol |
| UNII-CV0IB81ORO |
| CV0IB81ORO |
| EINECS 205-636-4 |
| INS NO.161H(I) |
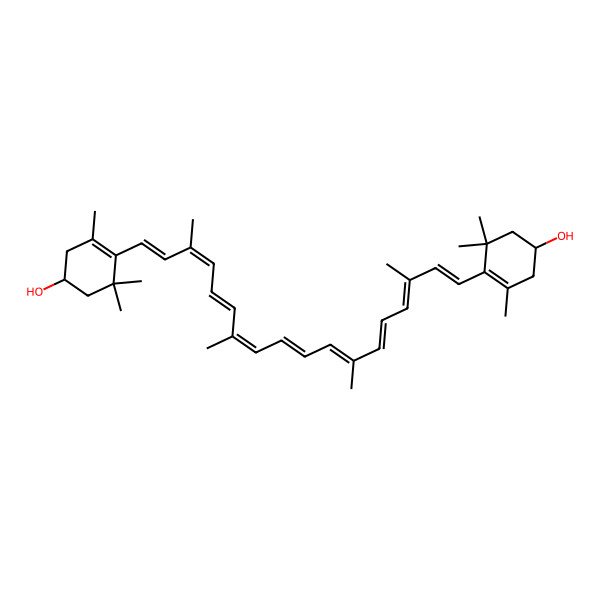
| (1R)-4-[(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4R)-4-hydroxy-2,6,6-trimethylcyclohexen-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethylcyclohex-3-en-1-ol |
| DTXSID5046807 |
| CHEBI:27547 |
| INS-161H(I) |
| all-trans-Anchovyxanthin |
| all-trans-beta-carotene-3,3'-diol |
| E 161H |
| E-161H(I) |
| (1R,1'R)-4,4'-((1E,3E,5E,7E,9E,11E,13E,15E,17E)-3,7,12,16-Tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaene-1,18-diyl)bis(3,5,5-trimethylcyclohex-3-enol) |
| DTXCID3026807 |
| MFCD08435940 |
| (3R,3'R)-dihydroxy-beta,beta-carotene |
| ZEAXANTHIN (MART.) |
| ZEAXANTHIN [MART.] |
| .beta.,.beta.-Carotene-3,3'-diol, (3R,3'R)- |
| Zeaxanthine |
| Zeaxanthins |
| 3R,3'R Zeaxanthin |
| Beta Carotene 3,3' Diol |
| Zeaxanthin, tech. |
| NCGC00181020-01 |
| (9Z)-Zeaxanthin |
| Zeaxanthin, 65% |
| (13Z)-Zeaxanthin |
| Zeaxanthin - 10% |
| 3R 3'R Zeaxanthin |
| Zeaxanthin, all-trans- |
| ZEAXANTHIN [MI] |
| ZEAXANTHIN [INCI] |
| ZEAXANTHIN [VANDF] |
| Anchovyxanthin, all-trans- |
| ss,ss-carotene-3,3'-diol |
| ZEAXANTHIN [WHO-DD] |
| SCHEMBL19442 |
| Zeaxanthin, analytical standard |
| CHEMBL2359248 |
| beta,beta-carotene-3R,3'R-diol |
| Tox21_112670 |
| LMPR01070261 |
| NSC713073 |
| DB11176 |
| DS-5859 |
| NSC-713073 |
| (3R,3'R)-DIHYDROXY-beta-CAROTENE |
| Zeaxanthin 10 microg/mL in Acetonitrile |
| CAS-144-68-3 |
| XZ177654 |
| HY-120318 |
| LS-185818 |
| CS-0077564 |
| (3R,3'R)-DIHYDROXY-.BETA.-CAROTENE |
| beta,beta-Carotene-3,3'-diol, (3R,3'R)- |
| C06098 |
| (3R,3'R,15Z)-beta,beta-Carotene-3,3'-diol |
| EN300-7418250 |
| A808258 |
| Q169337 |
| W-108152 |
| .beta.-Carotene-3,3'-diol, (3R,3'R)-all-trans- |
| beta,beta-carotene-3,3'-diol, (3R,3'R)-all-trans- |
| 6AB548E1-4B81-4843-8E0E-481DCFC93CA8 |
| ZEAXANTHIN/(3R,3'R)-ZEAXANTHIN (CONSTITUENT OF AZTEC MARIGOLD ZEAXANTHIN EXTRACT) |
| (1R)-3,5,5-trimethyl-4-[(1E,3E,5E,7E,9E,11E,13E,15E,17E)-3,7,12,16-tetramethyl-18-[(4R)-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol |
| (1R)-4-[(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4R)-4-hydroxy-2,6,6-trimethyl-1-cyclohexenyl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-1-cyclohex-3-enol |
| (1R)-4-[(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl]-3,5,5-trimethylcyclohex-3-en-1-ol |
| 4-(18-(4-hydroxy-2,6,6-trimethyl-1-cyclohexenyl)-3,7,12,16-tetramethyl-octadeca-1,3,5,7,9,11,13,15,17-nonaenyl)-3,5,5-trimethyl-cyclohex-3-en-1-ol |
|
There are more than 10 synonyms. If you wish to see them all click here.
|