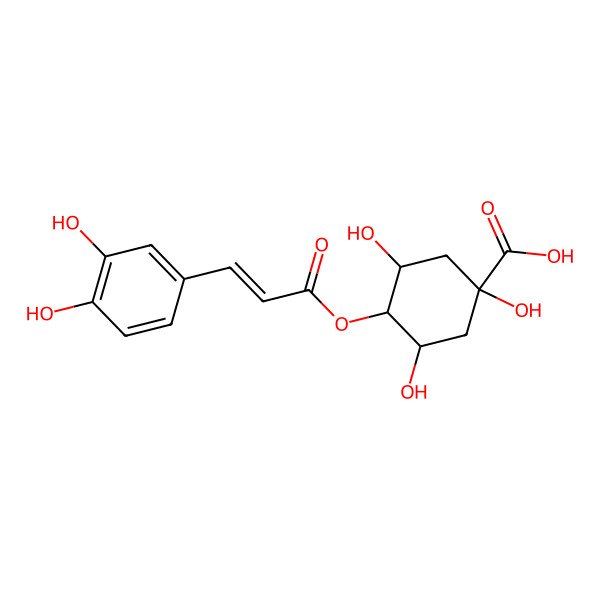
| 905-99-7 |
| 4-Caffeoylquinic acid |
| 4-O-Caffeoylquinic acid |
| 4-Cqa |
| 4-o-Caffeoyl quinic acid |
| Quinic acid 4-O-caffeate |
| 4-O-trans-caffeoylquinic acid |
| 4-O-(3,4-Dihydroxycinnamoyl)-D-quinic acid |
| F23DJ84IZ9 |
| (3R,5R)-4-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy-1,3,5-trihydroxycyclohexane-1-carboxylic acid |
| CHEMBL4203706 |
| 4-(3,4-Dihydroxycinnamoyl)quinic acid |
| (1alpha,3alpha,4alpha,5beta)-4-(3-(3,4-(Dihydroxyphenyl)-1-oxo-2-propenyl)oxy)-1,3,5-trihydroxycyclohexanecarboxylic acid |
| 82638-23-1 |
| 87099-73-8 |
| rel-(1S,3R,4S,5R)-4-(((E)-3-(3,4-Dihydroxyphenyl)acryloyl)oxy)-1,3,5-trihydroxycyclohexanecarboxylic acid |
| MFCD10566638 |
| Cryptochlorogenic-acid |
| 4-Caffeoylquinic acid;4-O-Caffeoylquinic acid |
| Quinic acid, 4-caffeoyl- |
| UNII-F23DJ84IZ9 |
| 4-O-(E)-caffeoylquinic acid |
| CHEMBL3092676 |
| Cinnamic acid, 3,4-dihydroxy-, 4-carboxy-2,4,6-trihydroxycyclohexyl ester |
| Quinic acid, 4-caffeoyl-, E- |
| SCHEMBL18180782 |
| SCHEMBL20883249 |
| ACon1_000120 |
| CHEBI:75491 |
| HY-N0787 |
| BDBM50455380 |
| s9319 |
| 4-O-Caffeoylquinic acid, >=98.0% |
| AKOS037514601 |
| CCG-268076 |
| CS-3767 |
| NCGC00180861-01 |
| (3R,5R)-4-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy-1,3,5-trihydroxy-cyclohexanecarboxylic acid |
| BS-17821 |
| PD118874 |
| Cryptochlorogenic acid, analytical standard |
| 4-Caffeoylquinic acid/ Cryptochlorogenic acid |
| Q-100886 |
| B0B55D52-5101-4E6D-AA67-72D1CECBAA5A |
| Q27145347 |
| 49B68A4C-6EC9-4441-B7D7-E3B740A8CDEC |
| (1alpha,3R,4alpha,5R)-4-[[3-(3,4-Dihydroxyphenyl)-1-oxo-2-propen-1-yl]oxy]-1,3,5-trihydroxycyclohexanecarboxylic acid |
| (1S,3R,4S,5R)-4-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}-1,3,5-trihydroxycyclohexanecarboxylic acid |
| 1,3beta,5alpha-Trihydroxy-4alpha-(3,4-dihydroxycinnamoyloxy)cyclohexane-1beta-carboxylic acid |
| CYCLOHEXANECARBOXYLIC ACID, 4-(((2E)-3-(3,4-DIHYDROXYPHENYL)-1-OXO-2-PROPEN-1-YL)OXY)-1,3,5-TRIHYDROXY-, (1.ALPHA.,3R,4.ALPHA.,5R)- |
| Cyclohexanecarboxylic acid, 4-(((2E)-3-(3,4-dihydroxyphenyl)-1-oxo-2-propen-1-yl)oxy)-1,3,5-trihydroxy-, (1alpha,3R,4alpha,5R)- |
| Cyclohexanecarboxylic acid, 4-((3-(3,4-dihydroxyphenyl)-1-oxo-2-1R-propenyl)oxy)-1,3,5-trihydroxy-, (1R-(1alpha,3alpha,4alpha,5beta))- |
| CYCLOHEXANECARBOXYLIC ACID, 4-((3-(3,4-DIHYDROXYPHENYL)-1-OXO-2-PROPEN-1-YL)OXY)-1,3,5-TRIHYDROXY-, (1.ALPHA.,3R,4.ALPHA.,5R)- |
| Cyclohexanecarboxylic acid, 4-((3-(3,4-dihydroxyphenyl)-1-oxo-2-propen-1-yl)oxy)-1,3,5-trihydroxy-, (1alpha,3R,4alpha,5R)- |
| Cyclohexanecarboxylic acid, 4-((3-(3,4-dihydroxyphenyl)-1-oxo-2-propenyl)oxy)-1,3,5-trihydroxy-, (1alpha,3R,4alpha,5R)- |
| Cyclohexanecarboxylic acid, 4-(3-(3,4-(dihydroxyphenyl)-1-oxo-2-propenyl)oxy)-1,3,5-trihydroxy-, (1alpha,3alpha,4alpha,5beta)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|