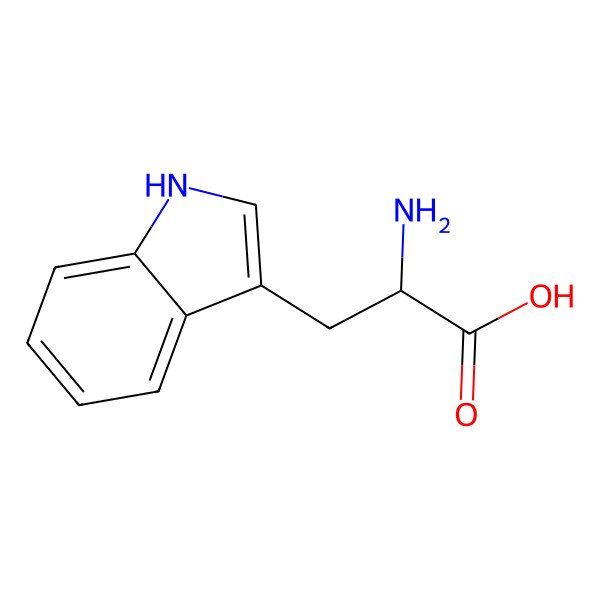
| tryptophan |
| 73-22-3 |
| L-Tryptophane |
| h-Trp-oh |
| (S)-Tryptophan |
| Tryptophane |
| trofan |
| tryptacin |
| Ardeytropin |
| Optimax |
| Pacitron |
| (2S)-2-amino-3-(1H-indol-3-yl)propanoic acid |
| Indole-3-alanine |
| Kalma |
| L-beta-3-Indolylalanine |
| L-Tryptofan |
| L-Trp |
| L-(-)-Tryptophan |
| 3-Indol-3-ylalanine |
| (-)-Tryptophan |
| Tryptophan (H-3) |
| 1-beta-3-Indolylalanine |
| triptofano |
| Tryptophanum |
| Tryptan |
| Lyphan |
| 1beta-3-Indolylalanine |
| Tryptophan (VAN) |
| 2-Amino-3-indolylpropanoic acid |
| Triptofano [Spanish] |
| Tryptophanum [Latin] |
| 1H-Indole-3-alanine |
| Tryptophan, L- |
| L(-)-Tryptophan |
| (L)-TRYPTOPHAN |
| (S)-alpha-Amino-1H-indole-3-propanoic acid |
| Tryptophane [French] |
| alpha'-Amino-3-indolepropionic acid |
| (S)-alpha-Aminoindole-3-propionic acid |
| Tryptophan [USAN:INN] |
| L-alpha-amino-3-indolepropionic acid |
| EH 121 |
| L-alpha-Aminoindole-3-propionic acid |
| trp |
| Sedanoct |
| Trytophan- |
| CCRIS 617 |
| 1H-Indole-3-alanine (VAN) |
| HSDB 4142 |
| Alanine, 3-indol-3-yl- |
| NCI-C01729 |
| (S)-alpha-amino-beta-(3-indolyl)-propionic acid |
| AI3-18478 |
| l-b-3-Indolylalanine |
| L-Alanine, 3-(1H-indol-3-yl)- |
| Indole-3-propionic acid, alpha-amino- |
| 1H-Indole-3-alanine, (S)- |
| alpha-Amino-3-indolepropionic acid, L- |
| CHEBI:16828 |
| UNII-8DUH1N11BX |
| (S)-2-Amino-3-(3-indolyl)propionic acid |
| EINECS 200-795-6 |
| 8DUH1N11BX |
| NSC 13119 |
| L-Ttp |
| 1H-Indole-3-propanoic acid, alpha-amino-, (S)- |
| L-(-)-Tryptophane |
| Propionic acid, 2-amino-3-indol-3-yl- |
| DTXSID5021419 |
| (S)-alpha-Amino-beta-indolepropionic acid |
| L-Tryptophan (9CI) |
| alpha-Amino-beta-(3-indolyl)-propionic acid |
| Tryptophan (USP/INN) |
| (S)-(-)-Tryptophan |
| (S)-a-Amino-b-indolepropionic acid |
| (S)-a-Aminoindole-3-propionic acid |
| Alanine, 3-indol-3-yl |
| Lopac-T-0254 |
| NSC-13119 |
| (S)-(-)-Tryptopha n |
| Tryptophan ((-),l,s) |
| (S)-a-Amino-1H-indole-3-propanoic acid |
| CHEMBL54976 |
| DTXCID801419 |
| TRP-01 |
| L-TRYPTOPHAN SIGMA GRADE |
| EC 200-795-6 |
| (S)-2-Amino-3-(1H-indol-3-yl)propanoic acid |
| alpha-Aminoindole-3-propionic acid |
| 2-amino-3-indol-3-ylpropionic acid |
| Tryptophanum (Latin) |
| Propionic acid, 2-amino-3-indol-3-yl |
| TRYPTOPHAN (II) |
| TRYPTOPHAN [II] |
| 2-Amino-3-(lH-indol-3-yl)-propanoic acid |
| S(-)-1-alpha-Aminoindole-3-propionic acid |
| MFCD00064340 |
| alpha-amino-beta-(3-indolyl)-pr opionic acid |
| T 0254 |
| TRYPTOPHAN (MART.) |
| TRYPTOPHAN [MART.] |
| (S)-(-)-2-Amino-3-(3-indolyl)propionic Acid |
| 80206-30-0 |
| Htrp |
| 151A3008-4CFE-40C9-AC0B-467EF0CB50EA |
| TRYPTOPHAN (EP MONOGRAPH) |
| TRYPTOPHAN [EP MONOGRAPH] |
| Levotryptophan |
| TRYPTOPHAN (USP MONOGRAPH) |
| TRYPTOPHAN [USP MONOGRAPH] |
| L-Tryptophan-13C11,15N2 |
| Alti-Tryptophan |
| 2-amino-3-indolylpropanic acid |
| beta-3-indolylalanine |
| D-Trp-OH |
| CAS-73-22-3 |
| N-ACETYLTRYPTOPHAN IMPURITY A (EP IMPURITY) |
| N-ACETYLTRYPTOPHAN IMPURITY A [EP IMPURITY] |
| (2S)-2-amino-3-(1H-indol-3-yl)propanoate |
| trytophan |
| l- tryptophan |
| L-Trytophan |
| 1qaw |
| l-a-Aminoindole-3-propionic acid |
| alpha-amino-beta-3-indolepropionic acid |
| L-Tryptophan,(S) |
| L-Trp-OH |
| Tryptophan [WHO-DD] |
| 2a4m |
| H-L-Trp-OH |
| TRYPTOPHAN [MI] |
| L-Tryptophan (JP15) |
| L-Tryptophan (JP17) |
| Tryptophan, L-(8CI) |
| TRYPTOPHAN [INN] |
| TRYPTOPHAN [HSDB] |
| TRYPTOPHAN [INCI] |
| TRYPTOPHAN [USAN] |
| TRP NH3+ COOH |
| Tryptophan (L-Tryptophan) |
| TRYPTOPHAN [VANDF] |
| Tryptophan, L- (8CI) |
| bmse000050 |
| bmse000868 |
| bmse001017 |
| D05EJG |
| Epitope ID:136043 |
| L-TRYPTOPHAN [FCC] |
| L-TRYPTOPHAN [JAN] |
| SCHEMBL7328 |
| 2-Amino-3-indolylpropanoate |
| (S)-1H-Indole-3-alanine |
| Lopac0_001183 |
| GTPL717 |
| L-TRYPTOPHAN [VANDF] |
| MLS001056750 |
| DivK1c_000457 |
| L-TRYPTOPHAN [USP-RS] |
| (s)-a-amino-b-indolepropionate |
| (S)-a-Aminoindole-3-propionate |
| BDBM21974 |
| HMS501G19 |
| KBio1_000457 |
| N06AX02 |
| 3-(1H-indol-3-yl)-L-Alanine |
| L-a-Amino-3-indolepropionic acid |
| NINDS_000457 |
| QIVBCDIJIAJPQS-VIFPVBQESA-N |
| HMS3263N07 |
| Pharmakon1600-01500600 |
| 202406-50-6 |
| HY-N0623 |
| STR02722 |
| (S)-alpha-Aminoindole-3-propionate |
| Tox21_201246 |
| Tox21_300359 |
| Tox21_501183 |
| NSC757373 |
| s3987 |
| (s)-alpha-amino-beta-indolepropionate |
| L-Tryptophan, Vetec(TM), 98.5% |
| (S)-a-Amino-1H-indole-3-propanoate |
| AKOS015854052 |
| Indoe-3-propionic acid, alpha-amino- |
| AM82273 |
| CCG-205257 |
| CS-W020011 |
| DB00150 |
| LP01183 |
| LS-1622 |
| NSC-757373 |
| SDCCGSBI-0051150.P002 |
| IDI1_000457 |
| NCGC00015994-01 |
| NCGC00094437-01 |
| NCGC00094437-02 |
| NCGC00094437-03 |
| NCGC00094437-04 |
| NCGC00094437-08 |
| NCGC00254424-01 |
| NCGC00258798-01 |
| NCGC00261868-01 |
| (S)-alpha-Amino-1H-indole-3-propanoate |
| AC-17050 |
| BP-13286 |
| IS_TRYPTOPHAN-2,4,5,6,7-D5 |
| SMR000326686 |
| TS-04426 |
| L-Tryptophan, BioUltra, >=99.5% (NT) |
| LS-185087 |
| EU-0101183 |
| T0541 |
| (S)-Tryptophan 1H-Indole-3-alanine, (S)- |
| EN300-52634 |
| C00078 |
| D00020 |
| L-.ALPHA.-AMINO-3-INDOLEPROPIONIC ACID |
| L-Tryptophan, reagent grade, >=98% (HPLC) |
| M02943 |
| P16427 |
| AB00373874_05 |
| L-Tryptophan, Vetec(TM) reagent grade, >=98% |
| (S)-2-amino-3-(1H-Indol-3-yl)-propionic acid |
| A837752 |
| L-Tryptophan, Cell Culture Reagent (H-L-Trp-OH) |
| Q181003 |
| SR-01000075590 |
| 4-(3-METHOXYANILINO)-4-OXOBUT-2-ENOICACID |
| SR-01000075590-1 |
| F0001-2364 |
| Z756440056 |
| 1H-INDOLE-3-PROPANOIC ACID, .ALPHA.-AMINO-, (S)- |
| L-Tryptophan, certified reference material, TraceCERT(R) |
| Tryptophan, European Pharmacopoeia (EP) Reference Standard |
| L-Tryptophan, United States Pharmacopeia (USP) Reference Standard |
| L-Tryptophan, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 99.0-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|