| 112-63-0 |
| Linoleic acid methyl ester |
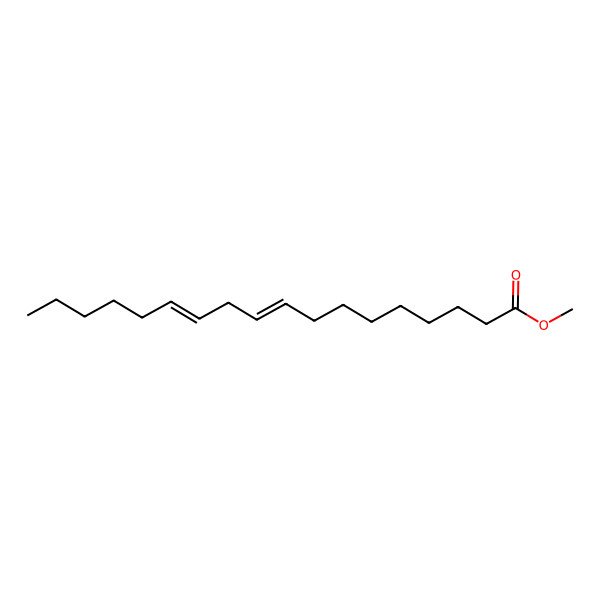
| methyl (9Z,12Z)-octadeca-9,12-dienoate |
| Methyl lineoleate |
| Linoleic acid, methyl ester |
| Methyl 9-cis,12-cis-octadecadienoate |
| Methyl octadecadienoate |
| Methyl linoleate, native |
| Methyl cis,cis-9,12-octadecadienoate |
| Linoleic acid,methyl ester |
| 9,12-Octadecadienoic acid (Z,Z)-, methyl ester |
| (9Z,12Z)-Methyl octadeca-9,12-dienoate |
| 9,12-Octadecadienoic acid (9Z,12Z)-, methyl ester |
| FEMA No. 3411 |
| EINECS 203-993-0 |
| linoleic acid-methyl ester |
| NSC 93981 |
| 68605-14-1 |
| 9,12-Octadecadienoic acid, methyl ester |
| cis-Linoleic acid methyl ester |
| AI3-03520 |
| DTXSID7020843 |
| UNII-24N6726DE5 |
| CHEBI:69080 |
| Methyl (Z,Z)-9,12-octadecadienoate |
| 24N6726DE5 |
| (Z,Z)-9,12-octadecadienoic acid methyl ester |
| 9,12-Octadecadienoic acid, methyl ester, (Z,Z) |
| cis-9,cis-12-Octadecadienoic acid, methyl ester |
| (9Z,12Z)-9,12-Octadecadienoic acid methyl ester |
| 9,12-Octadecadienoic acid, methyl ester, (Z,Z)- |
| DTXCID50843 |
| Methyl octadeca-9,12-dienoate |
| CAS-112-63-0 |
| methyl (9E,12Z)-octadeca-9,12-dienoate |
| Methyl linolate |
| MFCD00009534 |
| Natural methyl linoleate |
| LINOLEATE, METHYL |
| Telfairic Acid methyl ester |
| cis-9,cis-12-Octadecadienoic acid methyl ester |
| SCHEMBL56345 |
| METHYL LINOLEATE [MI] |
| RADIA 7062 |
| METHYL LINOLEATE [INCI] |
| CHEMBL3183866 |
| METHYL LINOLEATE [USP-RS] |
| Methyl linoleate, >=99% (GC) |
| Methyl (Z,Z)-9,12-octadienoate |
| HY-N1481 |
| EINECS 271-715-5 |
| Linoleic acid, methyl ester (8CI) |
| Tox21_201403 |
| Tox21_302917 |
| Methyl linoleate, analytical standard |
| NSC-93981 |
| s5759 |
| AKOS025310767 |
| METHYL OCTADEC-9,12-DIENOATE |
| LS-7503 |
| Methyl cis,cis-9, 12-octadecadienoate |
| 1-O-methyl-(9Z,12Z)-octadecadienoate |
| METHYL CIS-9,CIS-12 LINOLEATE |
| s12107 |
| NCGC00164324-01 |
| NCGC00164324-02 |
| NCGC00256520-01 |
| NCGC00258954-01 |
| (9Z,12Z)methyl octadeca-9,12-dienoate |
| AC-33782 |
| AS-63648 |
| (9Z,12Z)-Methyloctadeca-9,12-dienoate |
| Methyl (9Z,12Z)-9,12-octadecadienoate |
| FEMA NO. 3411, METHYL LINOLEATE- |
| (9Z,12Z)-Octadecadienoic acid methyl ester |
| CS-0017021 |
| L0078 |
| Methyl (9Z,12Z)-9,12-octadecadienoate # |
| METHYL CIS-9,CIS-12-OCTADECADIENOATE |
| Methyl ester(Z,Z)-9,12-Octadecadienoic acid |
| cis,cis-octadeca-9,12-dienoic acid methyl ester |
| Octadecadienoic acid methyl ester, 9,12-(Z,Z)- |
| (9Z,12Z)-octadeca-9,12-dienoic acid methyl ester |
| 9,12-Octadecadienoic acid, (Z,Z)-, methyl ester |
| J-002803 |
| Q27137420 |
| B7102443-24B7-4351-A24E-6617B1268CA6 |
| Methyl linoleate, United States Pharmacopeia (USP) Reference Standard |
|
There are more than 10 synonyms. If you wish to see them all click here.
|