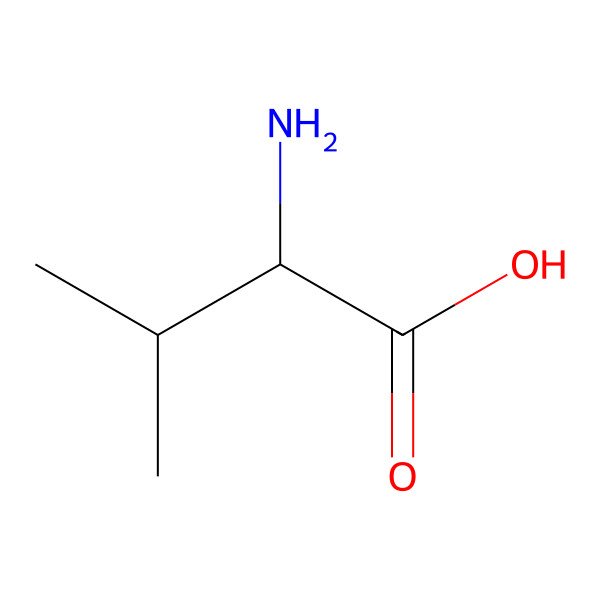
| valine |
| 72-18-4 |
| (S)-Valine |
| H-Val-OH |
| (S)-2-Amino-3-methylbutanoic acid |
| (2S)-2-amino-3-methylbutanoic acid |
| 2-Amino-3-methylbutyric acid |
| (S)-2-Amino-3-methylbutyric acid |
| L-alpha-Amino-beta-methylbutyric acid |
| L-valin |
| Valinum [Latin] |
| Valina [Spanish] |
| Butanoic acid, 2-amino-3-methyl- |
| valina |
| Valine (VAN) |
| Valinum |
| L(+)-alpha-Aminoisovaleric acid |
| (S)-alpha-Amino-beta-methylbutyric acid |
| Valine [USAN:INN] |
| Butanoic acid, 2-amino-3-methyl-, (S)- |
| 2-Amino-3-methylbutyric acid, (S)- |
| 2-Amino-3-methylbutanoic acid, (S)- |
| VALINE, L- |
| NSC 76038 |
| val |
| CHEBI:16414 |
| alpha-aminoisovaleric acid |
| l-(+)-alpha-Aminoisovaleric acid |
| UNII-HG18B9YRS7 |
| EINECS 200-773-6 |
| HG18B9YRS7 |
| NSC-76038 |
| (L)-valine |
| 2-Amino-3-methylbutanoic acid (VAN) |
| Valine (L-Valine) |
| HSDB 7800 |
| L-2-Aminoisovaleric Acid |
| L-Val |
| MFCD00064220 |
| 7004-03-7 |
| Valinum (Latin) |
| DTXSID40883233 |
| EC 200-773-6 |
| VALINE (II) |
| VALINE [II] |
| VALINE (MART.) |
| VALINE [MART.] |
| valin |
| Hval |
| VALINE (EP MONOGRAPH) |
| VALINE [EP MONOGRAPH] |
| VALINE (USP MONOGRAPH) |
| VALINE [USP MONOGRAPH] |
| l valine |
| (2S)-2-amino-3-methylbutanoate |
| L-2-Amino-3-methylbutanoic acid |
| LYSINE ACETATE IMPURITY D (EP IMPURITY) |
| LYSINE ACETATE IMPURITY D [EP IMPURITY] |
| 2-Amino-3-methyl-butyric acid |
| 2-amino-3-methylbutanoate |
| Racemic valine |
| L-(+)-a-Aminoisovaleric acid |
| s-valin |
| (S)-alpha-Aminoisovaleric acid |
| L-(+)-.alpha.-Aminoisovaleric acid |
| L-a-Amino-b-methylbutyric acid |
| L-valina |
| 3h-l-valine |
| (S)-a-Amino-b-methylbutyric acid |
| L-Valine; |
| L-Valine, FCC |
| Valine (USP) |
| (+)-valine |
| L-Valine,(S) |
| 1t4s |
| L-Valine (JP17) |
| L-Valine, 99% |
| VALINE [VANDF] |
| VALINE [HSDB] |
| VALINE [INCI] |
| VALINE [USAN] |
| (S)-Val |
| L-Val-4 |
| L-VAL-OH |
| VALINE [INN] |
| VALINE [WHO-DD] |
| VALINE [MI] |
| 2-Amino-3-methylbutyrate |
| L-VALINE [FCC] |
| L-VALINE [JAN] |
| bmse000052 |
| bmse000811 |
| bmse000860 |
| D0LL5V |
| L-(+)-a-Aminoisovalerate |
| L-a-Amino-b-methylbutyrate |
| SCHEMBL8516 |
| (S)-A-Aminoisovaleric acid |
| L-VALINE [USP-RS] |
| (S)-?-Aminoisovaleric acid |
| 2-Aminoisovaleric acid,(S) |
| CHEMBL43068 |
| (S)-a-Amino-b-methylbutyrate |
| L-(+)-alpha-Aminoisovalerate |
| L-iso-C3H7CH(NH2)COOH |
| GTPL4794 |
| (S)-2-Amino-3-methylbutyrate |
| (S)-2-Amino-3-methylbutanoate |
| L-alpha-Amino-beta-methylbutyrate |
| (S)-2-amino-3-methyl-Butanoate |
| DTXCID201022782 |
| Pharmakon1600-01301009 |
| HY-N0717 |
| (S)-alpha-Amino-beta-methylbutyrate |
| (S)-2-amino-3-methyl-butyric acid |
| BDBM50463208 |
| NSC760111 |
| s5628 |
| (S)-2-amino-3-methyl-Butanoic acid |
| L-Valine, 98.5-101.5% |
| AKOS015841564 |
| AM82363 |
| CCG-266067 |
| CS-W020706 |
| DB00161 |
| L-Valine, 99%, natural, FCC, FG |
| LS-2345 |
| NSC-760111 |
| NCGC00344520-01 |
| AS-12787 |
| L-Valine, BioUltra, >=99.5% (NT) |
| L-Valine, SAJ special grade, >=98.5% |
| L-Valine, reagent grade, >=98% (HPLC) |
| V0014 |
| EN300-52625 |
| L-Valine, Vetec(TM) reagent grade, >=98% |
| C00183 |
| D00039 |
| L-Valine, Cell Culture Reagent (H-L-Val-OH) |
| M02950 |
| V-1800 |
| (S)-(+)-2-AMINO-3-METHYLBUTYRIC ACID |
| Q483752 |
| Q-100919 |
| L-Valine, certified reference material, TraceCERT(R) |
| F8889-8698 |
| Valine, European Pharmacopoeia (EP) Reference Standard |
| Z756430564 |
| 1B39571B-0AE8-4A9A-AE80-4B898D11A981 |
| L-Valine, dimer, meets the analytical specifications of USP |
| L-Valine, United States Pharmacopeia (USP) Reference Standard |
| L-Valine, Pharmaceutical Secondary Standard; Certified Reference Material |
| InChI=1/C5H11NO2/c1-3(2)4(6)5(7)8/h3-4H,6H2,1-2H3,(H,7,8)/t4-/m0/s |
| L-Valine, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|