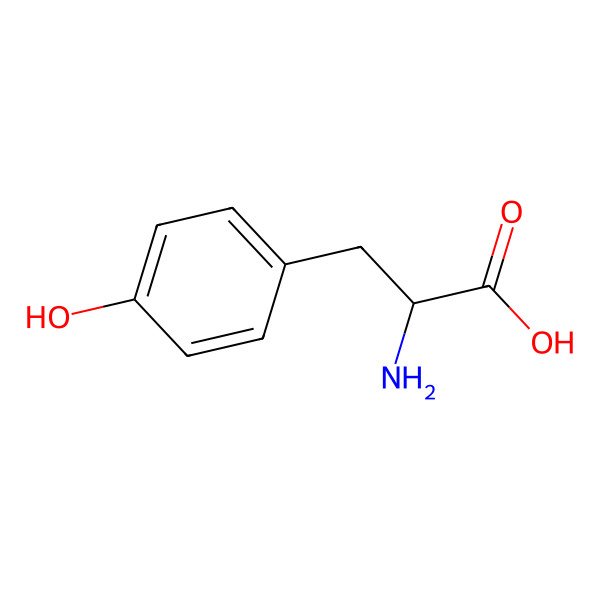
| tyrosine |
| 60-18-4 |
| (S)-Tyrosine |
| p-Tyrosine |
| L-p-Tyrosine |
| H-Tyr-OH |
| (2S)-2-amino-3-(4-hydroxyphenyl)propanoic acid |
| 4-Hydroxy-L-phenylalanine |
| Tyrosine, L- |
| Tyrosinum [Latin] |
| Tyrosine (VAN) |
| Tirosina [Spanish] |
| L-(-)-Tyrosine |
| (-)-alpha-Amino-p-hydroxyhydrocinnamic acid |
| L-Phenylalanine, 4-hydroxy- |
| beta-(p-Hydroxyphenyl)alanine |
| Tyrosine [USAN:INN] |
| FEMA No. 3736 |
| (S)-alpha-Amino-4-hydroxybenzenepropanoic acid |
| tirosina |
| Tyrosinum |
| L-2-Amino-3-p-hydroxyphenylpropanoic acid |
| 3-(4-Hydroxyphenyl)-L-alanine |
| (S)-3-(p-Hydroxyphenyl)alanine |
| (S)-(-)-Tyrosine |
| HSDB 2003 |
| alpha-Amino-beta-(4-hydroxyphenyl)propionic acid |
| L-Tyrosin |
| AI3-09055 |
| tyr |
| Propanoic acid, 2-amino-3-(4-hydroxyphenyl)-, (S)- |
| NSC 82624 |
| alpha-Amino-p-hydroxyhydrocinnamic acid, (-)- |
| L-Tyr |
| (S)-2-Amino-3-(4-hydroxyphenyl)propanoic acid |
| (S)-2-Amino-3-(p-hydroxyphenyl)propionic acid |
| L-Tyrosine, monomer |
| alpha-Amino-4-hydroxybenzenepropanoic acid, (S)- |
| 2-Amino-3-(4-hydroxyphenyl)propanoic acid, (S)- |
| L-Tyrosine (9CI) |
| Benzenepropanoic acid, alpha-amino-4-hydroxy-, (S)- |
| Tyrosine (USP/INN) |
| 4ts1 |
| EINECS 200-460-4 |
| MFCD00002606 |
| DTXSID1023730 |
| UNII-42HK56048U |
| CHEBI:17895 |
| (S)-2-Amino-3-(4-hydroxyphenyl)propionic acid |
| 3-(p-Hydroxyphenyl)alanine |
| NSC-82624 |
| L-Tyrosine (JAN) |
| 42HK56048U |
| NCGC00159350-02 |
| L-TYROSINE [JAN] |
| Melanin synthesized from Tyr substrate catalyzed by tyrosinase for 6 hrs |
| (-)-.alpha.-Amino-p-hydroxyhydrocinnamic acid |
| 2-Amino-3-(p-hydroxyphenyl)propionic acid |
| DTXCID603730 |
| 25619-78-7 |
| DD69927C-C6A8-4BC6-8E9A-0AB423B176E7 |
| Rxosine |
| Tyrosine Power |
| CAS-60-18-4 |
| Free-Form L-Tyrosine |
| -tyrosine |
| plovamer-acetate |
| Benzenepropanoate |
| 2csm |
| (L)-Tyrosine |
| (-) tyrosine |
| H-Tyr |
| L-Tyrosine,(S) |
| Tyrosine (L-Tyrosine) |
| L-Tyrosine (JP17) |
| TYROSINE [HSDB] |
| TYROSINE [INCI] |
| TYROSINE [USAN] |
| Tyrosine, L-(8CI) |
| TYROSINE [INN] |
| TYROSINE [II] |
| TYROSINE [MI] |
| L-TYR-OH |
| TYROSINE [VANDF] |
| Tyrosine, L- (8CI) |
| .alpha.-Amino-.beta.-(4-hydroxyphenyl)propionic acid |
| L-TYROSINE [FCC] |
| TYR NH3+ COOH |
| TYROSINE [MART.] |
| bmse000051 |
| CHEMBL925 |
| D01CRB |
| L-TYROSINE [FHFI] |
| TYROSINE [WHO-DD] |
| SCHEMBL1581 |
| L-[U-14C]Tyr |
| L-Phenylalanine-4-hydroxy- |
| L-Tyrosine non-animal source |
| L-TYROSINE [USP-RS] |
| Levodopa impurity, l-tyrosine- |
| TYROSINE [ORANGE BOOK] |
| CCRIS 6768 |
| GTPL4791 |
| L-Tyrosine, >=97%, FG |
| TYROSINE [EP MONOGRAPH] |
| TYROSINE [USP MONOGRAPH] |
| BDBM18129 |
| N-acetyl-o-(dihydroxymethylsilyl)- |
| HY-N0473 |
| L-Tyrosine, Vetec(TM), 98.5% |
| Tox21_111594 |
| (-)-a-Amino-p-hydroxyhydrocinnamate |
| AC2634 |
| s4608 |
| AKOS010400205 |
| IS_4-HYDROXYPHENYL-D4-ALANINE |
| Tox21_111594_1 |
| (C14-H19-N3-O5)x- |
| (S)-a-Amino-4-hydroxybenzenepropanoate |
| AM82304 |
| CS-8013 |
| CS-O-02423 |
| DB00135 |
| LS-2336 |
| (-)-alpha-Amino-p-hydroxyhydrocinnamate |
| (-)-a-Amino-p-hydroxyhydrocinnamic acid |
| (S)-3-(4-HYDROXYPHENYL)ALANINE |
| (S)-a-amino-4-hydroxy-Benzenepropanoate |
| NCGC00159350-03 |
| NCGC00344525-01 |
| AC-11295 |
| AS-11772 |
| BP-13285 |
| LEVODOPA IMPURITY B [EP IMPURITY] |
| (S)-alpha-Amino-4-hydroxybenzenepropanoate |
| DIETHYL1,3,5-BENZENETRICARBOXYLATE |
| L-Tyrosine, BioUltra, >=99.0% (NT) |
| (S)-2-Amino-3-(p-hydroxyphenyl)propionate |
| (S)-a-Amino-4-hydroxybenzenepropanoic acid |
| L-Tyrosine, Free Base - CAS 60-18-4 |
| (S)-a-amino-4-hydroxy-Benzenepropanoic acid |
| (S)-alpha-amino-4-hydroxy-Benzenepropanoate |
| L-Tyrosine, reagent grade, >=98% (HPLC) |
| L-Tyrosine, SAJ special grade, >=99.0% |
| T0550 |
| EN300-52629 |
| C00082 |
| D00022 |
| D70837 |
| L-Tyrosine, Vetec(TM) reagent grade, >=98% |
| M02963 |
| (2S)-2-amino-3-(4-hydroxyphenyl)propanoicacid |
| (S)-alpha-amino-4-hydroxy-Benzenepropanoic acid |
| L-Tyrosine, Cell Culture Reagent (H-L-Tyr-OH) |
| (S)-.alpha.-Amino-4-hydroxybenzenepropanoic acid |
| 2-Amino-3-(4-hydroxyphenyl)propanoic acid-(S)- |
| A832631 |
| N-ACETYLTYROSINE IMPURITY A [EP IMPURITY] |
| Q188017 |
| J-521656 |
| Propanoic acid, 2-amino-3-(4-hydroxyphenyl)-(S)- |
| LEVODOPA IMPURITY, L-TYROSINE- [USP IMPURITY] |
| Q27115106 |
| Benzenepropanoic acid, .alpha.-amino-4-hydroxy-, (S)- |
| F8889-8713 |
| L-Tyrosine, certified reference material, TraceCERT(R) |
| Tyrosine, European Pharmacopoeia (EP) Reference Standard |
| Z756440046 |
| L-Tyrosine, United States Pharmacopeia (USP) Reference Standard |
| 2-amino-3-(4-hydroxyphen yl)-2-amino-3-(4-hydroxyphenyl)-Propanoate |
| 2-amino-3-(4-hydroxyphen yl)-2-amino-3-(4-hydroxyphenyl)-Propanoic acid |
| Benzeneethanaminium,a-carboxy-4-hydroxy-N,N,N-trimethyl-,inner salt,(as)- |
| 1189756-47-5 |
| L-Tyrosine, from non-animal source, meets EP, USP testing specifications, suitable for cell culture, >=99.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|