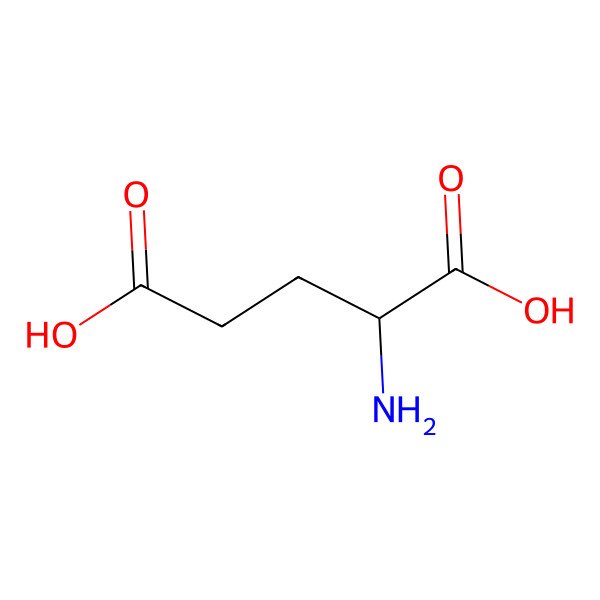
| GLUTAMIC ACID |
| 56-86-0 |
| (2S)-2-Aminopentanedioic acid |
| L-glutamate |
| (S)-2-Aminopentanedioic acid |
| Glutamidex |
| Glutaminol |
| H-Glu-OH |
| glutacid |
| Aciglut |
| glutaminic acid |
| L-Glutaminic acid |
| Glutamicol |
| Glutaton |
| (S)-Glutamic acid |
| Glusate |
| L-glu |
| L-(+)-glutamic acid |
| D-Glutamiensuur |
| alpha-aminoglutaric acid |
| Glutamic acid, L- |
| Acidum glutamicum |
| (S)-(+)-Glutamic acid |
| Acide glutamique |
| 2-Aminoglutaric acid |
| Acidum glutaminicum |
| L-2-Aminoglutaric acid |
| 1-Aminopropane-1,3-dicarboxylic acid |
| 25513-46-6 |
| Acido glutamico |
| FEMA No. 3285 |
| L-alpha-Aminoglutaric acid |
| Glutamic acid (H-3) |
| Glutamate, L- |
| alpha-Glutamic acid |
| glut |
| glutamate |
| Glutamic acid (VAN) |
| a-Glutamic acid |
| L-Glutaminsaeure |
| Glutamic acid, (S)- |
| Glutaminic acid (VAN) |
| CCRIS 7314 |
| L-Acido glutamico |
| Pentanedioic acid, 2-amino-, (S)- |
| glu |
| a-Aminoglutaric acid |
| Glutamic Acid [USAN:INN] |
| AI3-18472 |
| L-a-Aminoglutaric acid |
| Acide glutamique [INN-French] |
| Acido glutamico [INN-Spanish] |
| Acidum glutamicum [INN-Latin] |
| EPA Pesticide Chemical Code 374350 |
| NSC 143503 |
| alpha-Aminoglutaric acid (VAN) |
| Glutamic Acid (L-glutamic acid) |
| 2-Aminopentanedioic acid, (S)- |
| aminoglutaric acid |
| EINECS 200-293-7 |
| L-2-amino-pentanedioic acid |
| UNII-3KX376GY7L |
| 3KX376GY7L |
| INS NO.620 |
| DTXSID5020659 |
| CHEBI:16015 |
| Gamma-L-Glutamic Acid |
| INS-620 |
| L-Glutamic acid (9CI) |
| C5H9NO4 |
| NSC-143503 |
| L(+)-Glutamic acid |
| E620 |
| Glutamic acid, L-, peptides |
| DTXCID30659 |
| HSDB 490 |
| E 620 |
| E-620 |
| EC 200-293-7 |
| Sodium Glutamate (L-glutamic Acid) |
| NCGC00024502-03 |
| L-Glutamic acid (JAN) |
| (S)-2-AMINO-1,5-PENTANEDIOIC ACID |
| L(+)-Monosodium glutamate monohydrate |
| L-Glutamic acid-13C5 |
| L-GLUTAMIC ACID [JAN] |
| Acido glutamico (INN-Spanish) |
| Acidum glutamicum (INN-Latin) |
| (2S)-2-aminopentanedioate |
| l glutamic acid |
| GLUTAMIC ACID (EP MONOGRAPH) |
| GLUTAMIC ACID [EP MONOGRAPH] |
| .alpha.-Glutamic acid |
| ALANINE IMPURITY B (EP IMPURITY) |
| ALANINE IMPURITY B [EP IMPURITY] |
| 6899-05-4 |
| glt |
| 2-Amino-pentanedioic acid |
| LYSINE ACETATE IMPURITY B (EP IMPURITY) |
| LYSINE ACETATE IMPURITY B [EP IMPURITY] |
| 1-amino-propane-1,3-dicarboxylic acid |
| GLUTAMIC ACID [USAN] |
| 55443-55-5 |
| 6106-04-3 |
| MFCD00002634 |
| aminoglutarate |
| Gulutamine |
| alpha-Glutamate |
| a-Glutamate |
| L-gluatmate |
| a-Aminoglutarate |
| L-glutamic-acid |
| NSC143503 |
| L-Glutamic adid |
| 2-Aminoglutarate |
| Glutamate, L |
| 1ftj |
| 1xff |
| (S)-glutamate |
| Glutamic acid, L |
| L-a-Aminoglutarate |
| alpha-Aminoglutarate |
| Gulutamine (USP) |
| (L)-glutamic acid |
| H-Glu |
| L-Glutamic,(S) |
| L-(+)-Glutamate |
| L-alpha-Aminoglutarate |
| Glutamic acid (USP) |
| Tocris-0218 |
| [3h]-l-glutamic acid |
| 1ii5 |
| Polyglutamic acid(PGA) |
| (+)-L-Glutamic acid |
| (S)-(+)-Glutamate |
| (S)-Glu |
| L-[14C(U)]glutamate |
| (S)-2-Aminopentanedioate |
| Biomol-NT_000170 |
| D00ENY |
| GLUTAMIC ACID [MI] |
| L-Glutamic acid (JP17) |
| SCHEMBL2202 |
| GLUTAMIC ACID [INN] |
| L-Glutamic acid, 98.5% |
| Lopac0_000529 |
| S)-2-Aminopentanedioic acid |
| GLUTAMIC ACID [INCI] |
| GLUTAMIC ACID [VANDF] |
| L-GLUTAMIC ACID [FCC] |
| BPBio1_001132 |
| CHEMBL575060 |
| GTPL1369 |
| GLUTAMIC ACID [USP-RS] |
| GLUTAMIC ACID [WHO-DD] |
| L-GLUTAMIC ACID [FHFI] |
| L-Glutamic acid, 99%, FCC |
| BDBM17657 |
| CHEBI:53374 |
| Glutamic acid, L-(7CI,8CI) |
| 1-Aminopropane-1,3-dicarboxylate |
| (C5-H9-N-O4)x- |
| Glutamic acid, L- (7CI,8CI) |
| L (+)-glutamic acid, alpha-form |
| 1-amino-propane-1,3-dicarboxylate |
| 138-16-9 |
| L-Glutamic acid, non-animal source |
| Pentanedioic acid, 2-amino-, (S) |
| Tox21_113053 |
| HB0383 |
| HSCI1_000269 |
| PDSP1_000128 |
| PDSP1_001539 |
| PDSP2_000127 |
| PDSP2_001523 |
| s6266 |
| AKOS006238837 |
| AKOS015854087 |
| AM81690 |
| CCG-204619 |
| DB00142 |
| LS-2330 |
| SDCCGSBI-0050512.P002 |
| CAS-56-86-0 |
| NCGC00024502-01 |
| NCGC00024502-02 |
| NCGC00024502-04 |
| NCGC00024502-07 |
| (2S)-2-aminopentanedioic acid;H-Glu-OH |
| AC-11294 |
| DS-13284 |
| HY-14608 |
| LS-71885 |
| (S)-1-Aminopropane-1,3-dicarboxylic acid |
| (S)-1-Aminopropane-1,3-dicarboxylic acid |
| CS-0003473 |
| G0059 |
| EN300-52632 |
| L-Glutamic acid, BioUltra, >=99.5% (NT) |
| L-Glutamic acid, tested according to Ph.Eur. |
| C00025 |
| D00007 |
| L-Glutamic acid, NIST(R)RM 8573, USGS40 |
| M02979 |
| M03872 |
| Glutamic acid, L-; ((S)-(+)-Glutamic acid) |
| L-Glutamic acid, JIS special grade, >=99.0% |
| L-Glutamic acid, NIST(R) RM 8574, USGS41 |
| A831210 |
| Glutamic acid, L-; ((S)-(+)-Glutamic acid) |
| SR-01000597730 |
| J-502415 |
| L-Glutamic acid, ReagentPlus(R), >=99% (HPLC) |
| L-Glutamic acid, Vetec(TM) reagent grade, >=99% |
| SR-01000597730-1 |
| L-Glutamic acid, >=99%, FCC, natural sourced, FG |
| Q26995161 |
| F8889-8668 |
| Z756440052 |
| 27322E29-9696-49C1-B541-86BEF72DE2F3 |
| Glutamic acid, European Pharmacopoeia (EP) Reference Standard |
| L-Glutamic acid, certified reference material, TraceCERT(R) |
| Glutamic acid, United States Pharmacopeia (USP) Reference Standard |
| L-Glutamic acid, from non-animal source, meets EP testing specifications, suitable for cell culture, 98.5-100.5% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|