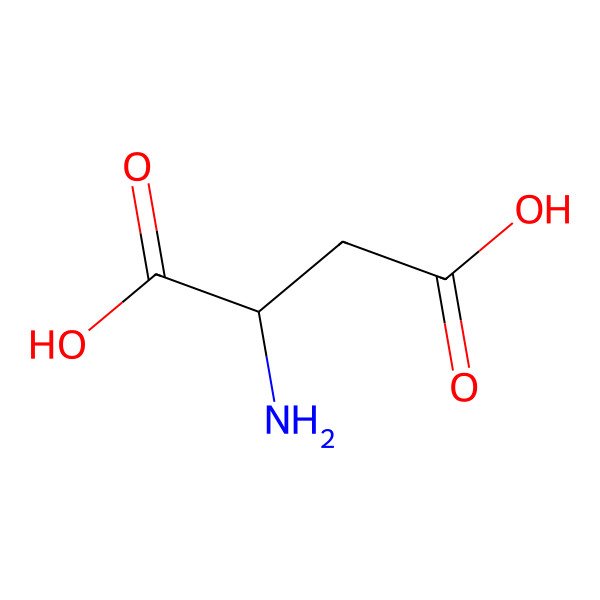
| aspartic acid |
| 56-84-8 |
| H-Asp-OH |
| aspartate |
| (2S)-2-aminobutanedioic acid |
| Asparagic acid |
| L-aspartate |
| Aspatofort |
| L-Aminosuccinic acid |
| (S)-2-Aminosuccinic acid |
| L-Asparagic acid |
| Asparaginic acid |
| L-Asparaginic acid |
| (S)-Aspartic acid |
| (2S)-Aspartic acid |
| (S)-Aminobutanedioic acid |
| Aspartic acid, L- |
| L-Aspartinsaeure |
| L-Asparaginsaeure |
| L-Asparaginsyra |
| Acidum asparticum |
| L-(+)-Aspartic acid |
| L-2-Aminobutanedioic acid |
| Aspartate, L- |
| Asparaginsaeure |
| L-Asp |
| 25608-40-6 |
| Aminosuccinic acid |
| Butanedioic acid, amino-, (S)- |
| Asparaginsaeure [German] |
| (+)-Aspartic acid |
| (L)-ASPARTIC ACID |
| Acido aspartico |
| Aspartic acid (VAN) |
| Acide aspartique |
| Asparagic acid (VAN) |
| (S)-(+)-Aminosuccinic acid |
| Asparaginic acid (VAN) |
| Acide aspartique [INN-French] |
| Acido aspartico [INN-Spanish] |
| HSDB 1430 |
| Deamidated asparagine |
| NSC 3973 |
| AI3-04461 |
| 2-aminosuccinic acid |
| FEMA No. 3565 |
| CCRIS 6181 |
| Acidum asparticum [INN-Latin] |
| 6899-03-2 |
| CHEBI:17053 |
| BRN 1723530 |
| UNII-30KYC7MIAI |
| 30KYC7MIAI |
| L( )-Aminobernsteinsaeure |
| L(+)-Aminobernsteinsaeure |
| asp |
| EINECS 200-291-6 |
| Aspartic Acid [USAN:INN] |
| (S)-2-aminobutanedioic acid |
| C4H7NO4 |
| Beta-L-Aspartic Acid |
| FEMA NO. 3656 |
| MFCD00002616 |
| (S)-(+)-Aspartic acid |
| 1-Amino-1,2-carboxyethane |
| CHEMBL274323 |
| DTXSID7022621 |
| Aspartic acid [USAN:USP:INN] |
| EC 200-291-6 |
| (S)-Aspartate |
| (+)-Aspartate |
| [3H]-L-aspartate |
| ASPARTIC ACID (II) |
| ASPARTIC ACID [II] |
| asparaginsaure |
| (S)-Aminobutanedioate |
| [3H]L-aspartic acid |
| [3H]-L-aspartic acid |
| ASPARTIC ACID (MART.) |
| ASPARTIC ACID [MART.] |
| ASPARTIC ACID (USP-RS) |
| ASPARTIC ACID [USP-RS] |
| NSC3973 |
| (2S)-2-azanylbutanedioic acid |
| ASPARTIC ACID (EP IMPURITY) |
| ASPARTIC ACID [EP IMPURITY] |
| L-Asparticacid |
| l aspartic acid |
| ASPARTIC ACID (EP MONOGRAPH) |
| ASPARTIC ACID [EP MONOGRAPH] |
| ASPARTIC ACID (USP MONOGRAPH) |
| ASPARTIC ACID [USP MONOGRAPH] |
| C4H7NO4.1/2Ca |
| ALANINE IMPURITY A (EP IMPURITY) |
| ALANINE IMPURITY A [EP IMPURITY] |
| (S)-Aminosuccinic Acid |
| C4-H7-N-O4.1/2Ca |
| LYSINE ACETATE IMPURITY A (EP IMPURITY) |
| LYSINE ACETATE IMPURITY A [EP IMPURITY] |
| 4-04-00-02998 (Beilstein Handbook Reference) |
| 55443-54-4 |
| 39162-75-9 |
| CHEBI:22660 |
| Aminosuccinate |
| Asparagate |
| Asparatate |
| NSC-3973 |
| aspartic acid,l |
| l-aspartic-acid |
| L-Asparagate |
| L-Aminosuccinate |
| aspartate magnesium |
| (L)-Aspartate |
| L- Aspartic acid |
| alpha-Aminosuccinate |
| (2S)-Aspartate |
| L-(+)-Aspartate |
| L-[14C]aspartate |
| L-Aspartic acid, 2 |
| (R)-2-aminosuccinate |
| (S)-2-aminosuccinate |
| Tocris-0214 |
| [3h]-l-asp |
| (S)-amino-Butanedioate |
| alpha-Aminosuccinic acid |
| (S)-(+)-Aspartate |
| L-Aspartic acid (9CI) |
| 2-Amino-3-methylsuccinate |
| Biomol-NT_000168 |
| bmse000031 |
| bmse000875 |
| D0OT9B |
| Aspartic acid (USP/INN) |
| ASPARTIC ACID [MI] |
| L-Aspartic acid (JP17) |
| SCHEMBL3231 |
| (S)-amino-Butanedioic acid |
| ASPARTIC ACID [INN] |
| L-Aspartic acid, >=98% |
| L-Aspartic acid, 99.0% |
| Lopac0_000133 |
| ASPARTIC ACID [HSDB] |
| ASPARTIC ACID [INCI] |
| ASPARTIC ACID [USAN] |
| Aspartic acid, L- (8CI) |
| ASPARTIC ACID [VANDF] |
| L-Aspartic acid (H-Asp-OH) |
| L-ASPARTIC ACID [FCC] |
| L-ASPARTIC ACID [JAN] |
| BPBio1_001128 |
| DTXCID402621 |
| GTPL3309 |
| GTPL4534 |
| ASPARTIC ACID [WHO-DD] |
| L-ASPARTIC ACID [FHFI] |
| aspartic acid (L-aspartic acid) |
| BDBM18125 |
| L-Aspartic acid, >=98%, FG |
| (C4-H7-N-O4)x- |
| HMS3260K08 |
| .alpha.-Aminosuccinic acid, (L)- |
| HY-N0666 |
| STR04614 |
| Tox21_500133 |
| HB0374 |
| PDSP1_000819 |
| PDSP2_000806 |
| AKOS006239578 |
| AKOS015853957 |
| AM81585 |
| CCG-204228 |
| DB00128 |
| LP00133 |
| LS-2569 |
| SDCCGSBI-0050121.P002 |
| potassium aspartate and magnesium aspart |
| L-Aspartic acid (H-Asp-OH) USP grade |
| NCGC00024499-01 |
| NCGC00024499-02 |
| NCGC00024499-03 |
| NCGC00024499-04 |
| NCGC00024499-05 |
| NCGC00024499-06 |
| NCGC00024499-10 |
| NCGC00260818-01 |
| BP-13291 |
| L-Aspartic acid, Vetec(TM) reagent grade |
| A0546 |
| CS-0009701 |
| EU-0100133 |
| L-Aspartic acid, BioXtra, >=99% (HPLC) |
| S5632 |
| EN300-64901 |
| L-Aspartic acid, BioUltra, >=99.5% (T) |
| A 9256 |
| A-9220 |
| C00049 |
| D00013 |
| D70832 |
| M03000 |
| L-Aspartic acid, SAJ special grade, >=99.0% |
| A817928 |
| A824434 |
| L-Aspartic acid, reagent grade, >=98% (HPLC) |
| Q178450 |
| SR-01000597734 |
| SR-01000597734-3 |
| F8889-8684 |
| Z995084132 |
| A4B5FB11-A4B6-4D75-9860-2ACF670700B9 |
| Aspartic acid, European Pharmacopoeia (EP) Reference Standard |
| L-Aspartic acid, certified reference material, TraceCERT(R) |
| Aspartic acid, United States Pharmacopeia (USP) Reference Standard |
| L-Aspartic acid, BioReagent, suitable for cell culture, suitable for insect cell culture |
| L-Aspartic acid, Pharmaceutical Secondary Standard; Certified Reference Material |
| 27881-03-4 |
| L-Aspartic acid, from non-animal source, meets EP, USP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|