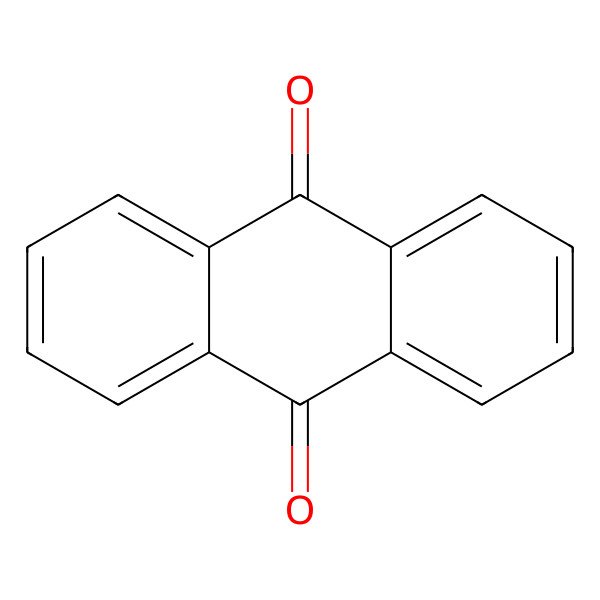
| 84-65-1 |
| anthracene-9,10-dione |
| 9,10-Anthraquinone |
| 9,10-Anthracenedione |
| Anthradione |
| 9,10-Dioxoanthracene |
| Hoelite |
| Morkit |
| Corbit |
| 9,10-Anthrachinon |
| Anthra-9,10-quinone |
| Anthrachinon |
| Anthrapel |
| 9,10-quinone |
| Az-Q |
| Avipel |
| Kawasaki saq |
| Caswell No. 052A |
| NSC 7957 |
| 9,10-Anthracendion |
| CCRIS 649 |
| DTXSID3020095 |
| CHEBI:40448 |
| HSDB 2074 |
| Anthracene, 9,10-dihydro-9,10-dioxo- |
| Anthracene-9,10-quinone |
| Anthraquinone [BSI:ISO] |
| UNII-030MS0JBDO |
| EINECS 201-549-0 |
| 030MS0JBDO |
| 9,10-Anthrachinon [Czech] |
| EPA Pesticide Chemical Code 122701 |
| FLIGHT CONTROL |
| AI3-09073 |
| NSC-7957 |
| 9,10-dihydro-9,10-dioxoanthracene |
| 9,10-dihydroanthracene-9,10-dione |
| DTXCID9095 |
| CHEMBL55659 |
| NSC7957 |
| EC 201-549-0 |
| NCGC00094960-03 |
| Anthraquinone 100 microg/mL in Acetonitrile |
| ANTHRAQUINONE (IARC) |
| ANTHRAQUINONE [IARC] |
| 1,4-bis-[2-(2-hydroxy-ethylamino)-ethylamino]-anthraquinone |
| CAS-84-65-1 |
| SAQ |
| dioxoanthracene |
| anthraquinon- |
| Anthracenequinone |
| anthracenequinones |
| (p) Anthrapel |
| 9,10-antraquinona |
| MFCD00001188 |
| 9,10-Anthraguinone |
| Anthraquinone, 97% |
| 9,10-Anthracendione |
| Spectrum_001527 |
| SpecPlus_000645 |
| Spectrum2_000405 |
| Spectrum3_001501 |
| Spectrum4_000907 |
| Spectrum5_001897 |
| an anthraquinone derivative |
| ANTHRAQUINONE [MI] |
| Anthra-9,10-quinone # |
| D0L5LN |
| Epitope ID:116191 |
| ANTHRAQUINONE [ISO] |
| ANTHRAQUINONE [HSDB] |
| ANTHRAQUINONE [INCI] |
| SCHEMBL14943 |
| BSPBio_003141 |
| KBioGR_001374 |
| KBioSS_002007 |
| DivK1c_006741 |
| SPECTRUM1502103 |
| SPBio_000330 |
| KBio1_001685 |
| KBio2_002007 |
| KBio2_004575 |
| KBio2_007143 |
| KBio3_002641 |
| Anthraquinone, analytical standard |
| HMS1921J14 |
| HY-N0354 |
| EINECS 292-087-9 |
| Tox21_111369 |
| BDBM50094892 |
| CCG-39966 |
| s5168 |
| STK398385 |
| Anthracene,10-dihydro-9,10-dioxo- |
| 9,10-Dihydro-9,10-diketoanthracene |
| AKOS000282964 |
| Tox21_111369_1 |
| DS-4810 |
| LS-1837 |
| USEPA/OPP Pesticide Code: 122701 |
| NCGC00094960-01 |
| NCGC00094960-02 |
| NCGC00094960-04 |
| NCGC00094960-06 |
| 90530-46-4 |
| AC-12719 |
| Anthraquinone, purum, >=99.0% (HPLC) |
| A0502 |
| CS-0008907 |
| FT-0622417 |
| Anthraquinone, Vetec(TM) reagent grade, 97% |
| EN300-71612 |
| A-8000 |
| Anthraquinone, PESTANAL(R), analytical standard |
| AB-131/40003277 |
| Q423174 |
| SR-01000944707 |
| SR-01000944707-1 |
| BRD-K15957397-001-03-4 |
| Melting point standard 283-286C, analytical standard |
| F0001-2133 |
| Z1142688857 |
| 71730-08-0 |
| 9,10-anthraquinone;9,10-anthracenedione;anthraquinone;anthracene-9,10-dione;9,10-anthraquinone 9,10-anthracenedione anthraquinone anthracene-9,10-dione |
| 9TA |
| InChI=1/C14H8O2/c15-13-9-5-1-2-6-10(9)14(16)12-8-4-3-7-11(12)13/h1-8 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|