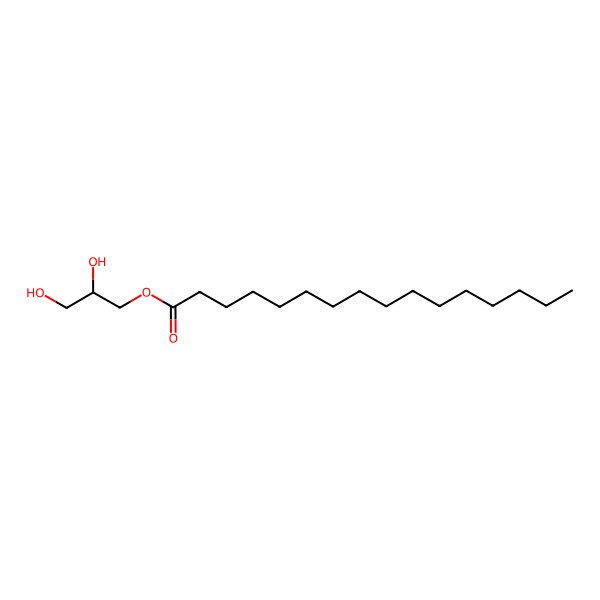
| Monopalmitin |
| 1-Monopalmitin |
| 1-Palmitoyl-rac-glycerol |
| GLYCERYL PALMITATE |
| 2,3-dihydroxypropyl hexadecanoate |
| 1-Palmitoylglycerol |
| Glycerol 1-monopalmitate |
| Glycerol 1-palmitate |
| Glycerol 3-palmitate |
| 19670-51-0 |
| 2,3-Dihydroxypropyl palmitate |
| 1-Monopalmitoylglycerol |
| Palmitin, 1-mono- |
| alpha-Monopalmitin |
| DL-alpha-Palmitin |
| Hexadecanoic acid, 2,3-dihydroxypropyl ester |
| 1-Monohexadecanoyl-rac-glycerol |
| .alpha.-Monopalmitin |
| L-alpha-Palmitin |
| Glycerides, C16-22 |
| Palmitoyl glycerol |
| 1-hexadecanoyl-rac-glycerol |
| Palmitic acid .alpha.-monoglyceride |
| glyceryl 1-palmitate |
| Glycerol monopalmitate |
| Palmitic acid alpha-monoglyceride |
| Glycerol palmitate |
| 1-Glycerol hexadecanoate |
| Glyceryl monopalmitate |
| NSC 404240 |
| 1-Glyceryl monohexadecanoate |
| UNII-U9H9OM3S75 |
| 26657-96-5 |
| MG(16:0/0:0/0:0)[rac] |
| U9H9OM3S75 |
| CHEBI:69081 |
| (S)-2,3-Dihydroxypropyl palmitate |
| EINECS 208-812-9 |
| EINECS 243-218-3 |
| EINECS 251-283-4 |
| EINECS 268-083-8 |
| (+-)-2,3-dihydroxypropyl palmitate |
| NSC404240 |
| NSC-404240 |
| (+-)-2,3-Dihydroxypropyl hexadecanoate |
| EC 268-083-8 |
| 68002-70-0 |
| Hexadecanoic acid, 2,3-dihydroxypropyl ester, (A+/-)- |
| Palmitin, mono- |
| C19H38O4 |
| 1-palmitoyl-glycerol |
| MG(16:0) |
| monopalmitin-1-glyceride |
| 1-Monohexadecanoylglycerol |
| glycerol 1-monohexadecanoate |
| rac-1-Palmitoylglycerol |
| 3-Palmitoyl-rac-glycerol |
| rac-Glycerol 1-palmitate |
| Hexadecanoic acid, monoester with 1,2,3-propanetriol |
| MG 16:0 |
| DL--Palmitin |
| Palmitin, 1-mono |
| (1)-2,3-Dihydroxypropyl palmitate |
| D,L-alpha-Palmitin |
| 1-hexadecanoylglycerol |
| rac-alpha-monopalmitin |
| rac-glyceryl palmitate |
| 1-Monopalmitate Glycerol |
| (+-)-alpha-monopalmitin |
| (+-)-glyceryl palmitate |
| DL-1-MONOPALMITIN |
| (+/-)-1-monopalmitin |
| Glycerides, C16-2-2 |
| Glycerol .alpha.-palmitate |
| rac-1-monopalmitoylglycerol |
| Glycerol alpha-Monopalmitate |
| rac-glycerol 1-monopalmitate |
| SCHEMBL74863 |
| (+-)--glycerol 1-palmitate |
| (+-)-1-monopalmitoylglycerol |
| DL-alpha-Palmitin, >=99% |
| (+-)-glycerol 1-monopalmitate |
| CHEMBL1078140 |
| CHEBI:75811 |
| DTXSID00891470 |
| rac-2,3-dihydroxypropyl palmitate |
| (.+/-.)-1-Hexadecanoylglycerol |
| AMY39945 |
| BCP30618 |
| MAG 16:0 |
| LMGL01010001 |
| MFCD00042734 |
| rac-palmitic acid alpha-monoglyceride |
| rac-2,3-dihydroxypropyl hexadecanoate |
| AKOS015902112 |
| (+/-)-1-HEXADECANOYLGLYCEROL |
| CCG-214232 |
| SB48400 |
| (+-)-palmitic acid alpha-monoglyceride |
| (+/-)-1-O-HEXADECANOYLGLYCEROL |
| Hexadecanoic acid,3-dihydroxypropyl ester |
| NCGC00186665-01 |
| AS-49753 |
| (.+/-.)-2,3-Dihydroxypropyl hexadecanoate |
| FT-0673487 |
| FT-0673489 |
| FT-0696902 |
| FT-0699567 |
| G0083 |
| L10008 |
| P-1125 |
| A870508 |
| SR-05000002594 |
| J-012718 |
| SR-05000002594-1 |
| 2,3-Dihydroxypropyl hexadecanoate(Chunks or pellets) |
| BRD-A80928489-001-01-0 |
| Q27887716 |
| 1-Monopalmitin;Glyceryl palmitate ;1-Palmitoyl-rac-glycerol;1-Palmitoylglycerol |
|
There are more than 10 synonyms. If you wish to see them all click here.
|