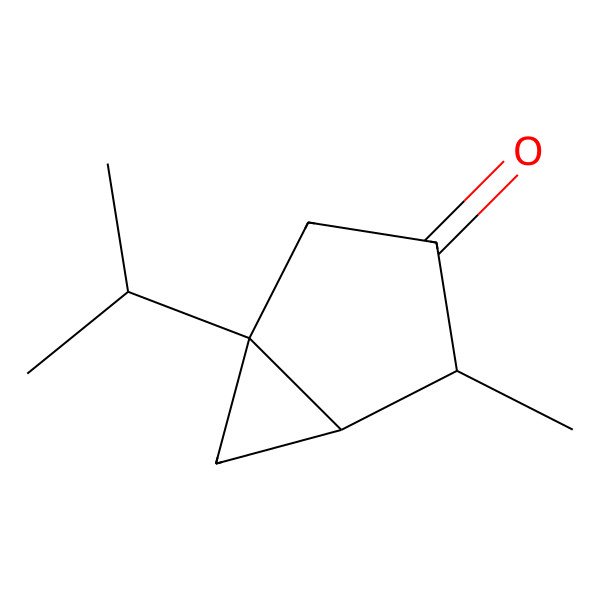
| 546-80-5 |
| alpha-(-)-Thujone |
| (-)-alpha-thujone |
| THUJONE |
| Thujon |
| l-Thujone |
| (-)-3-Isothujone |
| (-)-3-thujanone |
| Absinthol |
| dl-Thujone |
| 3-Thujanone, (-)- |
| (+-)-Isothujone |
| racemic 3-Isothujone |
| UNII-R0SQ9G0DU5 |
| EINECS 208-912-2 |
| NSC 93742 |
| BRN 4660369 |
| CCRIS 8582 |
| CHEBI:9577 |
| (1S,4R,5R)-thujan-3-one |
| (1S,4R,5R)-(-)-3-thujanone |
| 3-Thujanone, (+-)- |
| Isothujone, (-)- |
| R0SQ9G0DU5 |
| 3-Thujanone, (1S,4R,5R)-(-)- |
| 59573-80-7 |
| DTXSID3026148 |
| Bicyclo(3.1.0)hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1S,4R,5R)- |
| (-)-Thujone |
| (1S,4R,5R)-1-isopropyl-4-methylbicyclo[3.1.0]hexan-3-one |
| (1S,4R,5R)-4-methyl-1-(propan-2-yl)bicyclo[3.1.0]hexan-3-one |
| .alpha.-Thujone |
| Bicyclo[3.1.0]hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1S,4R,5R)- |
| Bicyclo(3.1.0)hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1-alpha,4-alpha,5-alpha)-(+-)- |
| 86HK1QRJ4K |
| CHEBI:50040 |
| (1S,4R,5R)-1-isopropyl-4-methylbicyclo(3.1.0)hexan-3-one |
| (1S,4R,5R)-4-methyl-1-(propan-2-yl)bicyclo(3.1.0)hexan-3-one |
| NSC-93742 |
| L-alpha-Thujone |
| (1S-(1alpha,4alpha,5alpha))-4-methyl-1-(1-methylethyl)bicyclo(3.1.0)hexan-3-one |
| [1S-(1alpha,4alpha,5alpha)]-4-methyl-1-(1-methylethyl)bicyclo[3.1.0]hexan-3-one |
| ??-Thujone |
| MFCD00001313 |
| (-)-trans-Thujone |
| (-)-a-Thujone |
| ALFA-THUJONE |
| (1S,4R)-1-Isopropyl-4-methylbicyclo[3.1.0]hexan-3-one |
| (-)- alpha -Thujone |
| (-)-alpha,beta-Thujone |
| a-(-)-THUJONE |
| D0TV0V |
| THUJONE, .ALPHA.- |
| MLS001065588 |
| SCHEMBL122447 |
| .ALPHA.-THUJONE [MI] |
| DTXCID206148 |
| GTPL5344 |
| CHEMBL1444078 |
| HMS3039B05 |
| (1R,2R,5S)-5-isopropyl-2-methyl-bicyclo[3.1.0]hexan-3-one |
| (1S,4R,5R)-4-methyl-1-propan-2-ylbicyclo[3.1.0]hexan-3-one |
| Bicyclo[3.1.0]hexan-3-one, 4-methyl-1-(1-methylethyl)-, [1S-(1.alpha.,4.alpha.,5.alpha.)]- |
| Tox21_200868 |
| (-)-Thujone ((-)-alpha-Thujone) |
| (1S,4R,5R)- 3-THUJANONE |
| AKOS040759330 |
| LMPR0102120019 |
| LS-2152 |
| NCGC00091438-01 |
| NCGC00091438-02 |
| NCGC00164360-01 |
| NCGC00258422-01 |
| CAS-546-80-5 |
| MS-22862 |
| SMR000568469 |
| (-)-alpha-Thujone, >=96.0% (GC) |
| HY-121618 |
| LS-153761 |
| CS-0082886 |
| C09906 |
| Q421838 |
| 1 - isopropyl - 4 - methylbicyclo(3.1.0)hexan - 3 - one |
| (-)-alpha-Thujone, (1S,4R)-1-Isopropyl-4-methylbicyclo[3.1.0]hexan-3-one |
| BICYCLO(3.1.0)HEXAN-3-ONE, 4-METHYL-1-(1-METHYLETHYL)-, (1S,5R)- |
| Bicyclo(3.1.0)hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1S-(1alpha,4alpha,5alpha))- |
| Bicyclo[3.1.0]hexan-3-one, 4-methyl-1-(1-methylethyl)-, [1S-(1?,4?,5?)]- |
| Bicyclo(3.1.0)hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1-alpha,4-alpha,5-alpha)-(+-)- (9CI) |
| Bicyclo[3.1.0]hexan-3-one, 4-methyl-1-(1-methylethyl)-, (1.alpha.,4.alpha.,5.alpha.)-(.+.)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|