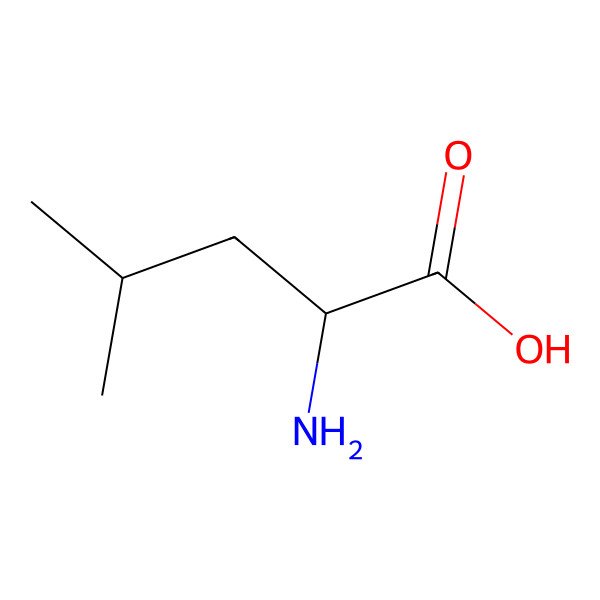
| leucine |
| 61-90-5 |
| (S)-2-Amino-4-methylpentanoic acid |
| (S)-Leucine |
| H-Leu-OH |
| (2S)-2-amino-4-methylpentanoic acid |
| Leucin |
| (S)-(+)-Leucine |
| L-Norvaline, 4-methyl- |
| LEUCINE, L- |
| (S)-2-Amino-4-methylvaleric acid |
| leu |
| L-alpha-Aminoisocaproic acid |
| Leucinum |
| FEMA No. 3297 |
| (2S)-alpha-leucine |
| L-leucin |
| Leucin [German] |
| L-(-)-2-Amino-4-methylpentanoic acid |
| alpha-Aminoisocaproic acid |
| Leucine (VAN) |
| Leucina |
| L-(+)-Leucine |
| Leucine [USAN:INN] |
| Leucinum [INN-Latin] |
| L-Leuzin |
| Leucina [INN-Spanish] |
| 2-amino-4-methylvaleric acid |
| Leucine, l |
| Valeric acid, 2-amino-4-methyl-, (S)- |
| Leucina [Latin,Spanish] |
| Pentanoic acid, 2-amino-4-methyl-, (S)- |
| 2-Amino-4-methylpentanoic acid, (S)- |
| 2-Amino-4-methylvaleric acid (L) |
| AI3-08899 |
| 2-Amino-4-methylpentanoic acid (L) |
| NSC 46709 |
| UNII-GMW67QNF9C |
| (2S)-alpha-2-amino-4-methylvaleric acid |
| GMW67QNF9C |
| EINECS 200-522-0 |
| NSC-46709 |
| L-Leu |
| Leucine (L-Leucine) |
| CHEBI:15603 |
| HSDB 7799 |
| alpha-Amino-gamma-methylvaleric acid |
| CHEMBL291962 |
| DTXSID9023203 |
| L-Leucine, labeled with tritium |
| EC 200-522-0 |
| L(+)-Leucine |
| Leucina (Latin,Spanish) |
| LEUCINE (II) |
| LEUCINE [II] |
| LEUCINE (MART.) |
| LEUCINE [MART.] |
| MFCD00002617 |
| Leuzin |
| Hleu |
| LEUCINE (EP MONOGRAPH) |
| LEUCINE [EP MONOGRAPH] |
| LEUCINE (USP MONOGRAPH) |
| LEUCINE [USP MONOGRAPH] |
| L-Isomer Leucine |
| VALINE IMPURITY C (EP IMPURITY) |
| VALINE IMPURITY C [EP IMPURITY] |
| Norvaline, 4-methyl- |
| 71000-80-1 |
| Leucine, L Isomer |
| Leucine, L-Isomer |
| (2S)-2-amino-4-methylpentanoate |
| L-a-Aminoisocaproic acid |
| 2-amino-4-methylpentanoicacid |
| NSC9252 |
| Valeric acid, 2-amino-4-methyl- |
| L-leucina |
| l- leucine |
| LeuOH |
| 1lan |
| 1usk |
| 3h-l-leucine |
| 4-methyl-norvalin |
| L-2-Amino-4-methylpentanoic acid |
| L-Leucine; |
| (L)-leucine |
| .alpha.-Amino-.gamma.-methylvaleric acid |
| (3H)Leucine |
| H-Leu |
| Leucine (USP) |
| L-a-Aminoisocaproate |
| 4-methyl-l-norvalin |
| L-Leucine,(S) |
| Leucine (H-3) |
| 4-methyl-L-Norvaline |
| L-Leu-OH |
| (2S)-2-amino-4-methyl-pentanoic acid |
| H-Leu-OH USP grade |
| 1f2o |
| starbld0005460 |
| L-Leucine (JP17) |
| L-alpha-Aminoisocaproate |
| LEUCINE [VANDF] |
| LEUCINE [HSDB] |
| LEUCINE [INCI] |
| LEUCINE [USAN] |
| LEUCINE [INN] |
| LEUCINE [MI] |
| L-LEUCINE [FCC] |
| L-LEUCINE [JAN] |
| 2-Aminoisobutylacetic acid |
| L-LEUCINE [FHFI] |
| LEUCINE [WHO-DD] |
| bmse000042 |
| bmse000920 |
| D0W4MZ |
| L-Leucine, 99%, FG |
| SCHEMBL3889 |
| NCIStruc1_001860 |
| NCIStruc2_000010 |
| iso-C4H9CH(NH2)COOH |
| 2-amino-4-methyl-valericaci |
| L-LEUCINE [USP-RS] |
| C6H12NO2T |
| 2-Amino-4-methyl-valeric acid |
| DTXCID903203 |
| GTPL3312 |
| L-.alpha.-Aminoisocaproic acid |
| (S)-2-Amino-4-methylvalerate |
| Norvaline, 4-methyl-, (L)- |
| 1,2-Amino-4-methylvaleric acid |
| (S)-2-Amino-4-methylpentanoate |
| (s)-2-amino-4-methylvalericacid |
| IS_LEUCINE-5,5,5-D3 |
| Pentanoic acid, 2-amino-4-methyl |
| Pharmakon1600-01301005 |
| C6-H12-N-O2-T |
| HY-N0486 |
| NCI46709 |
| STR01720 |
| L-Leucine, Vetec(TM), 98.5% |
| BDBM50219348 |
| CCG-37658 |
| NCGC00013565 |
| NSC760100 |
| s3753 |
| Oxirane, 2,3-bis(2-chlorophenyl)- |
| (S)-2-Amino-4-methyl-pentanoic acid |
| 2-Amino-4-methylvaleric acid, (L)- |
| AKOS010373766 |
| AKOS015841779 |
| AM81871 |
| CS-W020705 |
| DB00149 |
| NSC-760100 |
| 2-Amino-4-methylpentanoic acid, (L)- |
| L-Leucine, tested according to Ph.Eur. |
| NCGC00013565-02 |
| NCGC00096678-01 |
| Certified Reference Material of L-leucine |
| E641 |
| LS-87803 |
| 1-Leucine;2-Amino-4-methylpentanoic acid |
| L-Leucine, BioUltra, >=99.5% (NT) |
| L-Leucine, SAJ special grade, >=99.0% |
| BB 0256932 |
| L-Leucine, reagent grade, >=98% (HPLC) |
| L0029 |
| EN300-52627 |
| L-Leucine, Vetec(TM) reagent grade, >=98% |
| C00123 |
| D00030 |
| F70226 |
| L-2700 |
| M03060 |
| (2S)-2-azaniumyl-4-methyl-pentanoate;H-Leu-OH |
| L-Leucine, Cell Culture Reagent (H-L-Leu-OH) |
| A821449 |
| A833479 |
| Q483745 |
| Q-201312 |
| (2R)-2-amino-4-methyl-pentanoic acid;D-HOMO-VALINE |
| L-Leucine, certified reference material, TraceCERT(R) |
| LYSINE HYDROCHLORIDE IMPURITY A [EP IMPURITY] |
| 2B9FF792-3CA1-4BEA-BC63-6D4E1A86714E |
| F8889-8638 |
| Leucine, European Pharmacopoeia (EP) Reference Standard |
| Z756437526 |
| L-Leucine, United States Pharmacopeia (USP) Reference Standard |
| (S)-2-Amino-4-methyl-pentanoic acid methyl ester hydrochloride |
| L-Leucine, Pharmaceutical Secondary Standard; Certified Reference Material |
| 25248-98-0 |
| L-Leucine, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|