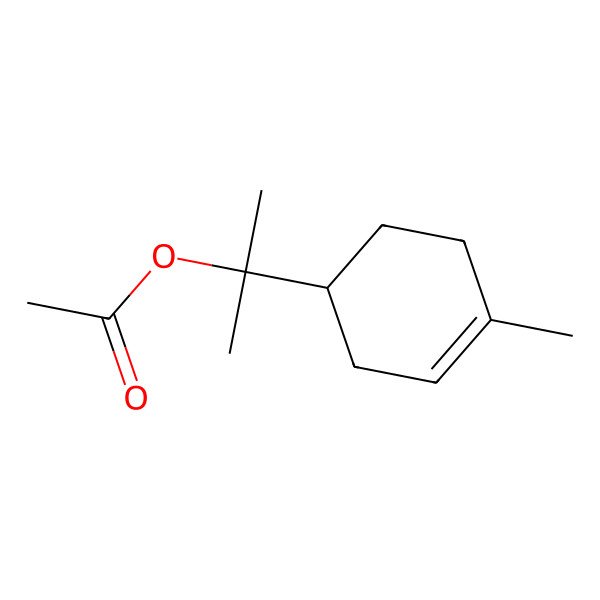
| ALPHA-TERPINYL ACETATE |
| 80-26-2 |
| Terpineol, acetate |
| 8007-35-0 |
| p-Menth-1-en-8-yl acetate |
| Terpineol acetate |
| p-Menth-1-en-8-ol, acetate |
| alpha-Terpineol acetate |
| (+/-)-alpha-Terpinyl acetate |
| alpha-Terpineol, acetate |
| .alpha.-Terpinyl acetate |
| 2-(4-methylcyclohex-3-en-1-yl)propan-2-yl acetate |
| UNII-NIT9SZT3D7 |
| .alpha.-Terpineol acetate |
| Terpinyl acetate (natural) |
| UNII-9RXE0I9F2J |
| 9RXE0I9F2J |
| FEMA No. 3047 |
| DTXSID2026496 |
| CHEBI:32320 |
| 3-Cyclohexene-1-methanol, .alpha.,.alpha.,4-trimethyl-, acetate |
| EINECS 201-265-7 |
| EINECS 232-357-5 |
| EINECS 234-183-5 |
| MFCD00037155 |
| (+/-)-2-(4-Methyl-3-cyclohexenyl)isopropyl acetate |
| BRN 3198769 |
| 3-Cyclohexene-1-methanol, alpha,alpha,4-trimethyl-, acetate |
| AI3-00522 |
| EC 201-265-7 |
| 4-06-00-00253 (Beilstein Handbook Reference) |
| DTXCID506496 |
| alpha,alpha,4-Trimethyl-3-cyclohexene-1-methyl acetate |
| 3-Cyclohexene-1-methanol, .alpha.,.alpha.,4-trimethyl-, 1-acetate |
| CAS-80-26-2 |
| d-\a-Terpineol acetate |
| 10581-37-0 |
| 1-methyl-1-(4-methylcyclohex-3-en-1-yl)ethyl acetate |
| lindenyl acetate |
| -erpinyl acetate |
| 3-Cyclohexene-1-methanol, .alpha.,.alpha.,4-trimethyl-, acetate, (1R)- |
| p-Menth-1-en-8-ol, acetate, ()- |
| terpinenyl acetate |
| a-Terpineol acetate |
| a-?Terpinyl acetate |
| para-menthenol acetate |
| NIT9SZT3D7 |
| Terpinyl acetate; >85% |
| 1-Methyl-1-(4-methyl-3-cyclohexen-1-yl)ethyl acetate # |
| 3-Cyclohexene-1-methanol, ?,?,4-trimethyl-, acetate, (R)- |
| SCHEMBL580624 |
| (R) - a,a,4 - trimethylcyclohex - 3 - ene - 1 - methyl acetate |
| CHEMBL3183581 |
| FEMA 3047 |
| [1-methyl-1-(4-methylcyclohex-3-en-1-yl)ethyl] acetate |
| ACETIC ACID TERPINYL ESTER |
| IGODOXYLBBXFDW-UHFFFAOYSA-N |
| alpha-Terpinyl acetate, (+/-)- |
| .ALPHA.-TERPINEOL, ACETATE |
| HY-N7136 |
| Tox21_202188 |
| Tox21_303242 |
| s5691 |
| alpha-Terpinyl acetate, >=95%, FG |
| AKOS015964845 |
| LMPR0102090045 |
| NCGC00164334-01 |
| NCGC00164334-02 |
| NCGC00256977-01 |
| NCGC00259737-01 |
| MS-23062 |
| p - menth - 1 - en - 8 - yl acetate |
| .ALPHA.-TERPINYL ACETATE, (+/-)- |
| .ALPHA.-TERPINYL ACETATE. (+/-)- |
| CS-0099572 |
| FT-0622203 |
| FT-0693969 |
| T0023 |
| E80789 |
| MENTHEN-1-YL-8-PROPIONATE;FEMA 3047 |
| 2-(4-methylcyclohex-3-enyl)propan-2-yl acetate |
| 2-(4-methyl-1-cyclohex-3-enyl)propan-2-yl acetate |
| Q4456134 |
| 1-Methyl-1-(4-methylcyclohex-3-enyl)ethyl ethanoate |
| 3-Cyclohexene-1-methanol, ?,?,4-trimethyl-, acetate |
| (+/-)-ALPHA-TERPINYL ACETATE;ALPHA-TERPINYL ACETATE |
| 2-(4-METHYL-3-CYCLOHEXEN-1-YL)-2-PROPYL ACETATE |
| (+-)-alpha,alpha,4-trimethylcyclohex-3-ene-1-methyl acetate |
| 3-Cyclohexene-1-methanol, alpha,alpha,4-trimethyl-, 1-acetate |
| .ALPHA.,.ALPHA.,4-TRIMETHYL-3-CYCLOHEXENE-1-METHYL ACETATE |
| 3-CYCLOHEXENE-1-METHANOL, ALPHA, ALPHA, 4-TRIMETHYL:ACETATE |
| (+/-)-alpha-Terpinyl acetate, predominantly alpha-isomer, analytical standard |
| (+/-)-alpha-Terpinyl acetate, predominantly alpha-isomer, technical, >=90% (GC) |
|
There are more than 10 synonyms. If you wish to see them all click here.
|