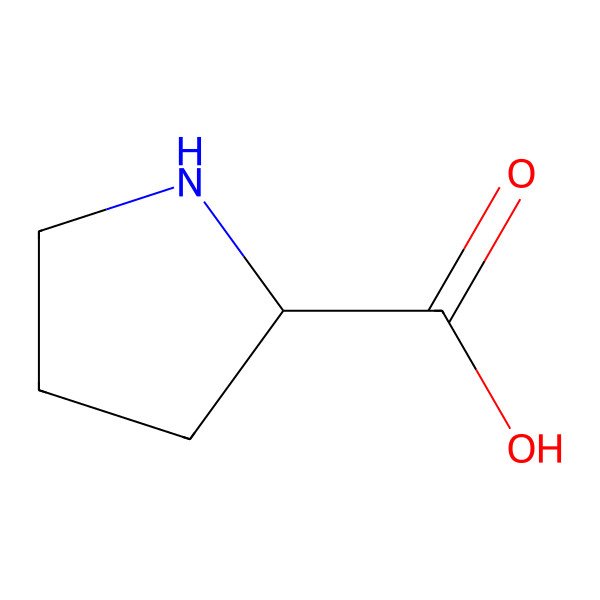
| proline |
| 147-85-3 |
| L-(-)-Proline |
| (S)-Pyrrolidine-2-carboxylic acid |
| (2S)-pyrrolidine-2-carboxylic acid |
| H-Pro-OH |
| 2-pyrrolidinecarboxylic acid |
| (-)-Proline |
| (-)-(S)-Proline |
| Prolinum |
| (S)-2-Pyrrolidinecarboxylic acid |
| (-)-2-Pyrrolidinecarboxylic acid |
| prolina |
| (S)-Proline |
| L-Pyrrolidine-2-carboxylic acid |
| L-alpha-Pyrrolidinecarboxylic acid |
| (S)-2-Carboxypyrrolidine |
| L-Prolin |
| Prolinum [Latin] |
| Prolina [Spanish] |
| Proline, L- |
| Proline (VAN) |
| (L)-PROLINE |
| FEMA No. 3319 |
| Proline [USAN:INN] |
| FEMA Number 3319 |
| 2-Pyrrolidinecarboxylic acid, (S)- |
| PRO (IUPAC abbreviation) |
| CB 1707 |
| (S)-(-)-Proline |
| HSDB 1210 |
| AI3-26710 |
| UNII-9DLQ4CIU6V |
| 9DLQ4CIU6V |
| Carboxypyrrolidine |
| L-(2,3-3H)Proline |
| EINECS 205-702-2 |
| NSC 46703 |
| Proline (USP) |
| pro |
| CHEBI:17203 |
| L-Proline, labeled with carbon-14 |
| NSC-46703 |
| CHEMBL54922 |
| L-2-pyrrolidinecarboxylic acid |
| 37159-97-0 |
| DTXSID5044021 |
| EC 205-702-2 |
| Prolinum (Latin) |
| prol |
| L-Proline (JAN) |
| Proline (L-Proline) |
| (-)-Proline (S)-2-Carboxypyrrolidine |
| PROLINE (II) |
| PROLINE [II] |
| L-PROLINE [JAN] |
| PROLINE (MART.) |
| PROLINE [MART.] |
| MFCD00064318 |
| 4305-67-3 |
| PROLINE (EP MONOGRAPH) |
| PROLINE [EP MONOGRAPH] |
| PROLINE (USP MONOGRAPH) |
| PROLINE [USP MONOGRAPH] |
| (2S)-pyrrolidin-1-ium-2-carboxylate |
| 2-Pyrrolidinecarboxylate |
| racemic proline |
| rac-proline |
| NSC46703 |
| s-proline |
| l- proline |
| 3h-l-proline |
| L-Proline; |
| (s)-prolin |
| femanumber3319 |
| H-Pro |
| (2S)-proline |
| Pro-OH |
| L-Proline,(S) |
| L-Pro-OH |
| L-Proline (JP17) |
| PROLINE [VANDF] |
| PROLINE [HSDB] |
| PROLINE [INCI] |
| PROLINE [USAN] |
| PROLINE [INN] |
| PROLINE [MI] |
| L-PROLINE [FCC] |
| PRO (IUPACabbreviation) |
| L-PROLINE [FHFI] |
| PROLINE [WHO-DD] |
| bmse000047 |
| bmse000947 |
| D0DZ3X |
| (2S)-2-carboxypyrrolidine |
| SCHEMBL7792 |
| L-PROLINE [USP-RS] |
| (S)-2-Pyrralidinecarboxylate |
| (S)-2-Pyrrolidinecarboxylate |
| (-)-2-Pyrrolidinecarboxylate |
| (s)-2-pyrrolidinecarboxylicaci |
| (S)-(-)-PROLIN |
| GTPL3314 |
| (s)-2-pyrrolidinecarboxylicacid |
| DTXCID3024021 |
| L-Proline, 99%, FCC, FG |
| (S)-2-Pyrralidinecarboxylic acid |
| pyrrolidin-2-(S)-carboxylic acid |
| Pharmakon1600-01301007 |
| (2S)-Pyrrolidin-2-carbonsalphaure |
| pyrrolidine-2-(S)-carboxylic acid |
| BCP25292 |
| HY-Y0252 |
| (S) -pyrrolidine-2-carboxylic acid |
| L-Proline, >=99.0% (NT) |
| (S)-(-)-Pyrrolidine-2-carboxylate |
| BDBM50000100 |
| NCGC00014017 |
| NSC760114 |
| s5629 |
| AKOS010372120 |
| AKOS015856025 |
| CCG-214709 |
| CS-W019861 |
| DB00172 |
| LS-2381 |
| NSC-760114 |
| (S)-(-)-Pyrrolidine-2-carboxylic acid |
| NCGC00014017-02 |
| NCGC00014017-03 |
| NCGC00097126-01 |
| AC-11190 |
| AS-10803 |
| L-Proline, BioUltra, >=99.5% (NT) |
| L-Proline, SAJ special grade, >=99.0% |
| AM20080359 |
| BB 0242381 |
| L-Proline, Vetec(TM), 98.5-101.5% |
| P0481 |
| EN300-52624 |
| L-Proline, Vetec(TM) reagent grade, >=99% |
| C00148 |
| D00035 |
| L-Proline, ReagentPlus(R), >=99% (HPLC) |
| M02947 |
| P17692 |
| Q-201327 |
| L-Proline, certified reference material, TraceCERT(R) |
| Q20035886 |
| A01B5B63-CC3D-4796-A7B4-C2DE26A6FA93 |
| F0001-2348 |
| Flavor and Extract Manufacturers' Association No. 3319 |
| Proline, European Pharmacopoeia (EP) Reference Standard |
| Z756429958 |
| L-Proline, United States Pharmacopeia (USP) Reference Standard |
| L-Proline, Pharmaceutical Secondary Standard; Certified Reference Material |
| L-Proline, from non-animal source, meets EP, USP testing specifications, suitable for cell culture |
|
There are more than 10 synonyms. If you wish to see them all click here.
|