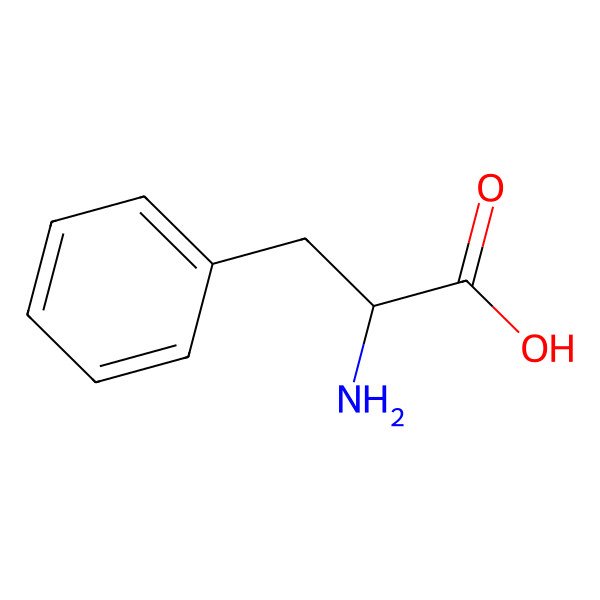
| phenylalanine |
| 63-91-2 |
| (2S)-2-amino-3-phenylpropanoic acid |
| 3-Phenyl-L-alanine |
| (S)-2-Amino-3-phenylpropanoic acid |
| (S)-Phenylalanine |
| (S)-2-Amino-3-phenylpropionic acid |
| (L)-Phenylalanine |
| 3-Phenylalanine |
| beta-Phenyl-L-alanine |
| Antibiotic FN 1636 |
| Alanine, 3-phenyl- |
| H-Phe-OH |
| L-Alanine, phenyl- |
| (S)-alpha-Amino-beta-phenylpropionic acid |
| L-Antibiotic FN 1636 |
| fenilalanina |
| Phenylalaninum |
| L-Alanine, 3-phenyl- |
| (S)-alpha-Aminohydrocinnamic acid |
| (S)-(-)-Phenylalanine |
| FEMA No. 3585 |
| Phenylalanine (VAN) |
| beta-Phenylalnine, (-)- |
| (S)-alpha-Amino-benzenepropanoic acid |
| Fenilalanina [Spanish] |
| Phenylalaninum [Latin] |
| Alanine, phenyl-, L- |
| endophenyl |
| Hydrocinnamic acid, alpha-amino- |
| Phenylalanine, L- |
| HSDB 1825 |
| laevo-phenyl alanine |
| alpha-Aminohydrocinnamic acid |
| phe |
| Benzenepropanoic acid, alpha-amino-, (S)- |
| 2-Amino-3-phenylpropionic acid, L- |
| NSC 79477 |
| Phenylalamine |
| Phenylalanine [USAN:INN:JAN] |
| alpha-Aminohydrocinnamic acid, L- |
| alpha-Amino-beta-phenylpropionic acid, L- |
| Alanine, phenyl- |
| EINECS 200-568-1 |
| (-)-phenylalanine |
| L-Phe |
| UNII-47E5O17Y3R |
| CCRIS 4254 |
| L-PHENYLALININE |
| CHEBI:17295 |
| 47E5O17Y3R |
| (-)-beta-Phenylalanine |
| beta-Phenyl-alpha-alanine |
| alpha-Amino-beta-phenylpropionic acid |
| NSC-79477 |
| 67675-33-6 |
| Phenylalanine (USP/INN) |
| phenylalanin |
| CHEMBL301523 |
| DTXSID4040763 |
| NCI9959 |
| (S)-alpha-Aminobenzenepropanoic acid |
| Phenylalanine [USAN:USP:INN:JAN] |
| Phenylalaninum (Latin) |
| MFCD00064227 |
| PHENYLALANINE (II) |
| PHENYLALANINE [II] |
| 2S-alpha-phenylalanine |
| PHENYLALANINE (MART.) |
| PHENYLALANINE [MART.] |
| Phenylalanine (L-Phenylalanine) |
| 1F9436B3-8B0D-4AC6-A004-4249B0BDA436 |
| PHENYLALANINE (EP MONOGRAPH) |
| PHENYLALANINE [EP MONOGRAPH] |
| PHENYLALANINE (USP MONOGRAPH) |
| PHENYLALANINE [USP MONOGRAPH] |
| .beta.-Phenylalanine |
| LEUCINE IMPURITY C (EP IMPURITY) |
| LEUCINE IMPURITY C [EP IMPURITY] |
| Phenyl-.alpha.-alanine |
| L-.beta.-Phenylalanine |
| TYROSINE IMPURITY A (EP IMPURITY) |
| TYROSINE IMPURITY A [EP IMPURITY] |
| .beta.-Phenyl-L-alanine |
| NATEGLINIDE IMPURITY D (EP IMPURITY) |
| NATEGLINIDE IMPURITY D [EP IMPURITY] |
| (-)-.beta.-Phenylalanine |
| .alpha.-Aminohydrocinnamic acid |
| .beta.-Phenyl-.alpha.-alanine |
| L-phenylaniline |
| Hydrocinnamic acid, .alpha.-amino- |
| 1-phenylalanine |
| .beta.-Phenyl-.alpha.-alanine, l- |
| L-phenyl Alanine |
| PheOH |
| 1usi |
| .alpha.-Amino-.beta.-phenylpropionic acid |
| racemic phenylalanine |
| Phenyl-alpha-alanine |
| 1f2p |
| alpha-Aminohydrocinnamate |
| L-Phenylalanine, 99% |
| 3- phenyl- L- alanine |
| L-Phenylalanine (JP15) |
| L-Phenylalanine (JP17) |
| PHENYLALANINE [MI] |
| bmse000045 |
| bmse000900 |
| bmse000921 |
| bmse001016 |
| D0R1CR |
| PHENYLALANINE [INN] |
| SCHEMBL8119 |
| NCIStruc1_000204 |
| NCIStruc2_000248 |
| PHENYLALANINE [HSDB] |
| PHENYLALANINE [INCI] |
| PHENYLALANINE [USAN] |
| PHENYLALANINE [VANDF] |
| (S)-alpha-Aminohydrocinnamate |
| L-Phenylalanine (H-Phe-OH) |
| L-PHENYLALANINE [FCC] |
| L-PHENYLALANINE [JAN] |
| L-2-Amino-3-phenylpropionate |
| L-PHENYLALANINE [FHFI] |
| L-Phenylalanine, 99%, FCC |
| PHENYLALANINE [WHO-DD] |
| GTPL3313 |
| (S)-alpha-Aminobenzenepropanoate |
| DTXCID2020763 |
| (S)-2-amino-3-phenylpropanoate |
| BDBM18073 |
| (S)-2-Amino-3-phenylpropionate |
| 2-Amino-3-phenyl-pronanoic acid |
| L-PHENYLALANINE [USP-RS] |
| L-Phenylalanine non-animal source |
| (S)-alpha-Amino-benzenepropanoate |
| L-2-Amino-3-phenylpropionic acid |
| HY-N0215 |
| L-2-amino-3-phenyl-propionic acid |
| AC8117 |
| CCG-37572 |
| NCGC00013103 |
| (S)-alpha-Aminiobenzenepropanoic acid |
| L-[2,3,4,5,6-3H]phenylalanine |
| (S)-alpha-Amino-beta-phenylpropionate |
| AKOS010373257 |
| AKOS015853585 |
| (S)-.alpha.-Aminobenzenepropanoic acid |
| DB00120 |
| L-Phenylalanine, Vetec(TM), 98.5% |
| LS-1515 |
| L-Phenylalanine, reagent grade, >=98% |
| NCGC00013103-02 |
| NCGC00013103-03 |
| NCGC00013103-04 |
| NCGC00013103-05 |
| NCGC00095047-01 |
| NCGC00095047-02 |
| NCGC00095047-03 |
| NCGC00095047-04 |
| AC-22417 |
| AS-14129 |
| BP-20538 |
| IS_PHENYLALANINE-2,3,4,5,6-D5 |
| L-Phenylalanine, 99%, natural, FCC, FG |
| AM20060774 |
| P0134 |
| Benzenepropanoic acid, .alpha.-amino-, (S)- |
| EN300-52626 |
| L-Phenylalanine, BioUltra, >=99.0% (NT) |
| A20654 |
| C00079 |
| D00021 |
| M02961 |
| L-Phenylalanine, SAJ special grade, >=99.0% |
| Q170545 |
| L-.ALPHA.-AMINO-.BETA.-PHENYLPROPIONIC ACID |
| L-Phenylalanine, Vetec(TM) reagent grade, >=98% |
| Q-201326 |
| L-Phenylalanine, Cell Culture Reagent (H-L-Phe-OH) |
| LYSINE HYDROCHLORIDE IMPURITY B [EP IMPURITY] |
| F0001-2360 |
| Z756430566 |
| L-Phenylalanine, certified reference material, TraceCERT(R) |
| L-PHENYL ALANINE (SEE ALSO 22839-47-0, ASPARTAME |
| L-Phenylalanine, 4-(bis(2-chloroethyl)amino)-N-formyl-(9CI) |
| Phenylalanine, European Pharmacopoeia (EP) Reference Standard |
| 3-(2-AMINOETHYL)-1,3-THIAZOLIDINE-2,4-DIONEHYDROCHLORIDE |
| L-Phenylalanine, United States Pharmacopeia (USP) Reference Standard |
| L(-)-phenylalanine; Beta-phenylalanine;Dl-2-amino-3-phenylpropanoic acid; |
| L-Phenylalanine, analytical standard, for Nitrogen Determination According to Kjeldahl Method |
| L-Phenylalanine, Pharmaceutical Secondary Standard; Certified Reference Material |
| L-PHENYL ALANINE (SEE ALSO 22839-47-0, ASPARTAME; 303-47-9 OCHRATOXINA; 673-06-3 D-PHENYL) |
| L-Phenylalanine, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|