| 26544-34-3 |
| Apigenin-7-apioglucoside |
| Apioside |
| UNII-6QU3EZE37U |
| 6QU3EZE37U |
| CHEBI:15932 |
| EINECS 247-780-0 |
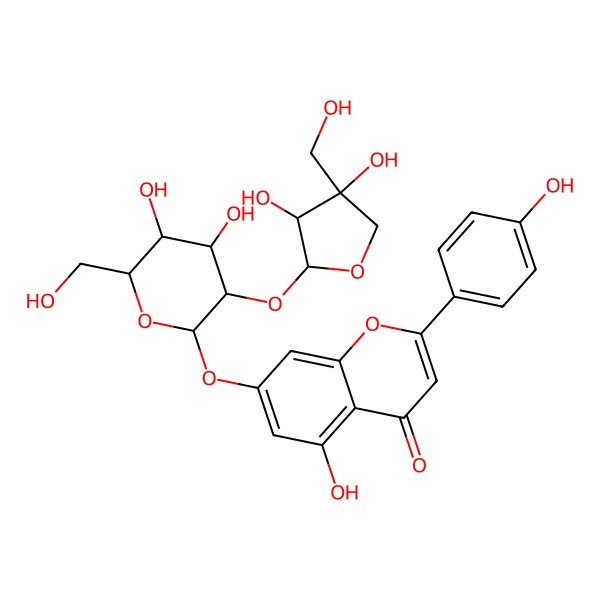
| 7-((2-O-beta-D-Apiofuranosyl-beta-D-glucopyranosyl)oxy)-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyranone |
| Apigenin 7-O-[beta-D-apiosyl-(1->2)-beta-D-glucoside] |
| 7-O-(beta-D-Apiofuranosyl-1,2-beta-D-glucosyl)-5,7,4'-trihydroxyflavone |
| 5-hydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl 3-C-(hydroxymethyl)-beta-D-glycero-tetrofuranosyl-(1->2)-beta-D-glucopyranoside |
| 7-(((2S,3R,4S,5S,6R)-3-(((2S,3R,4R)-3,4-Dihydroxy-4-(hydroxymethyl)tetrahydrofuran-2-yl)oxy)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)-5-hydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one |
| 7-[(2S,3R,4S,5S,6R)-3-[(2S,3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-5-hydroxy-2-(4-hydroxyphenyl)chromen-4-one |
| 5,7,4'-trihydroxyflavone 7-O-[beta-D-apiosyl-(1->2)-beta-D-glucoside] |
| 5-hydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl 2-O-[(2S,3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)tetrahydrofuran-2-yl]-beta-D-glucopyranoside |
| 7-[(2-O-D-Apio-beta-D-furanosyl-beta-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxy-phenyl)-4H-1-benzopyran-4-one |
| apigenin 7-O-(beta-D-apiosyl-(1->2)-beta-D-glucoside) |
| C26H28O14 |
| 5,7,4'-trihydroxyflavone 7-O-(beta-D-apiosyl-(1->2)-beta-D-glucoside) |
| 5-hydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl 2-O-((2S,3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)tetrahydrofuran-2-yl)-beta-D-glucopyranoside |
| 7-((2-O-D-Apio-beta-D-furanosyl-beta-D-glucopyranosyl)oxy)-5-hydroxy-2-(4-hydroxy-phenyl)-4H-1-benzopyran-4-one |
| 7-((2-O-D-Apio-beta-D-furanosyl-beta-D-glucopyranosyl)oxy)-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| 7-[(2-O-D-apio-beta-D-furanosyl-beta-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| Spectrum_000204 |
| SpecPlus_000939 |
| Apiin (11) |
| Spectrum2_001800 |
| Spectrum3_001787 |
| Spectrum4_001817 |
| Spectrum5_000553 |
| APIIN [MI] |
| BSPBio_003313 |
| KBioGR_002458 |
| KBioSS_000684 |
| SPECTRUM350025 |
| 7-O-(beta-D-apiofuranosyl-(1-2)-beta-D-glucosyl)-apigenin |
| MLS000575008 |
| DivK1c_007035 |
| SCHEMBL316910 |
| SPBio_001759 |
| CHEMBL1535342 |
| KBio1_001979 |
| KBio2_000684 |
| KBio2_003252 |
| KBio2_005820 |
| KBio3_002815 |
| DTXSID90949393 |
| 7-O-(beta-D-apiofuranosyl-(1-2)-beta-D-glucosyl)-5,7,4'-trihydroxyflavone |
| NTDLXWMIWOECHG-YRCFQSNFSA-N |
| BDBM153268 |
| HMS2209P05 |
| 4H-1-Benzopyran-4-one, 7-[(2-O-D-apio-.beta.-D-furanosyl-.beta.-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)- |
| HY-N0577 |
| CCG-39950 |
| AKOS030632988 |
| SDCCGMLS-0066949.P001 |
| NCGC00178147-01 |
| AS-74876 |
| SMR000156232 |
| CS-0009113 |
| C04858 |
| SR-01000712126 |
| Q2858300 |
| SR-01000712126-2 |
| BRD-K52379519-001-02-0 |
| 4H-1-Benzopyran-4-one, 7-[(2-O-D-apio-beta-D-furanosyl-beta-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)- |
| 7-((2-O-.BETA.-D-APIOFURANOSYL-.BETA.-D-GLUCOPYRANOSYL)OXY)-5- HYDROXY-2-(4-HYDROXYPHENYL)-4H-1-BENZOPYRANONE |
| 7-((2-O-D-APIO-.BETA.-D-FURANOSYL-.BETA.-D-GLUCOPYRANOSYL)OXY)- 5-HYDROXY-2-(4-HYDROXYPHENYL)-4H-1-BENZOPYRAN-4-ONE |
| 7-{[(2S,3R,4S,5S,6R)-3-{[(2S,3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy}-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-5-hydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one |
|
There are more than 10 synonyms. If you wish to see them all click here.
|