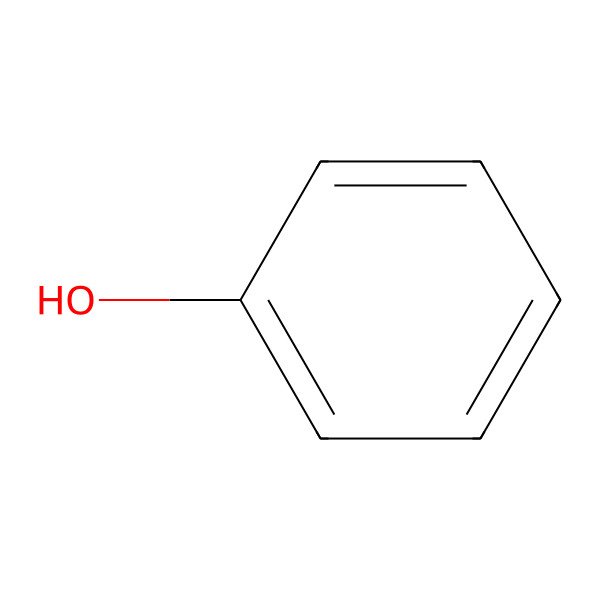
| 108-95-2 |
| carbolic acid |
| Hydroxybenzene |
| Phenic acid |
| Oxybenzene |
| Phenylic acid |
| Phenylic alcohol |
| Benzenol |
| Monophenol |
| Phenyl hydrate |
| Phenyl hydroxide |
| PhOH |
| Monohydroxybenzene |
| Phenyl alcohol |
| Paoscle |
| Phenole |
| Izal |
| Phenol alcohol |
| Acide carbolique |
| Phenol, liquefied |
| Fenolo |
| Benzene, hydroxy- |
| Carbolsaure |
| Phenosmolin |
| Fenol |
| Liquid phenol |
| Phenol, pure |
| Rcra waste number U188 |
| Liquefied phenol |
| Liquified Phenol |
| Carbolicum acidum |
| Fenolo [Italian] |
| NCI-C50124 |
| Phenole [German] |
| Baker's P & S liquid & Ointment |
| Campho-Phenique Gel |
| phenylalcohol |
| UN 2312 (molten) |
| Phenol [JAN] |
| Fenosmoline |
| UN 1671 (solid) |
| Baker's P and S Liquid and Ointment |
| Fenosmolin |
| Phenic |
| Carbolsaure [German] |
| Caswell No. 649 |
| Campho-Phenique Liquid |
| Phenol, molten |
| 2-allphenol |
| Fenol [Dutch, Polish] |
| NSC 36808 |
| Phenol, liquified |
| Monohydroxy benzene |
| Acide carbolique [French] |
| CCRIS 504 |
| Campho-Phenique Cold Sore Gel |
| Carbolsaeure |
| FEMA No. 3223 |
| Karbolsaeure |
| HSDB 113 |
| acide phenique |
| hydroxy benzene |
| DTXSID5021124 |
| Phenic alcohol |
| Phenol,liquified |
| Synthetic phenol |
| Phenol, liquid |
| Phenol, dimer |
| Phenol, solid |
| AI3-01814 |
| RCRA waste no. U188 |
| Baker's p and s |
| EINECS 203-632-7 |
| Phenol, sulfurated |
| UNII-339NCG44TV |
| MFCD00002143 |
| NSC-36808 |
| UN1671 |
| UN2312 |
| UN2821 |
| EPA Pesticide Chemical Code 064001 |
| 339NCG44TV |
| (14C)Phenol |
| CHEBI:15882 |
| Phenol [USP:JAN] |
| ENT-1814 |
| 27073-41-2 |
| CHEMBL14060 |
| DTXCID501124 |
| Phenol, labeled with carbon-14 |
| EC 203-632-7 |
| NSC36808 |
| 63496-48-0 |
| Phenol, solid [UN1671] [Poison] |
| Phenol, molten [UN2312] [Poison] |
| NCGC00091454-04 |
| PHENOL (IARC) |
| PHENOL [IARC] |
| PHENOL (II) |
| PHENOL [II] |
| PHENOL (MART.) |
| PHENOL [MART.] |
| Phenol, >=99.0% |
| 17442-59-0 |
| 65996-83-0 |
| PHENOL (EP MONOGRAPH) |
| PHENOL [EP MONOGRAPH] |
| PHENOL (USP MONOGRAPH) |
| PHENOL [USP MONOGRAPH] |
| Carbol |
| Phenol 100 microg/mL in Methanol |
| 61788-41-8 |
| 73607-76-8 |
| CAS-108-95-2 |
| METACRESOL IMPURITY A (EP IMPURITY) |
| METACRESOL IMPURITY A [EP IMPURITY] |
| SALICYLIC ACID IMPURITY C (EP IMPURITY) |
| SALICYLIC ACID IMPURITY C [EP IMPURITY] |
| HEXYLRESORCINOL IMPURITY A (EP IMPURITY) |
| HEXYLRESORCINOL IMPURITY A [EP IMPURITY] |
| PHENOL (2,3,4,5,6-D5) |
| arenols |
| Benzophenol |
| Karbolsaure |
| Hydroxy-benzene |
| Phnol |
| Phenol liquid |
| Phenol molten |
| Phenol synthetic |
| Pandy's reagent |
| Cepastat lozenges |
| Phenol (liquid) |
| 2-phenyl alcohol |
| Phenol, synthetic |
| Phenol, ultrapure |
| Phenol ACS grade |
| Paoscle (TN) |
| Carbolic acid liquid |
| Phenol (TN) |
| Phenol,(S) |
| Phenol, ACS reagent |
| Carbolic acid, liquid |
| PHENOL REAGENT |
| OXY BENZENE |
| PHENOL REAGE NT |
| 1ai7 |
| 1li2 |
| 4i7l |
| Liquefied phenol (TN) |
| CBO (CHRIS Code) |
| PHN (CHRIS Code) |
| PHENOL [VANDF] |
| PHENOL [FHFI] |
| PHENOL [HSDB] |
| PHENOL [INCI] |
| Phenol (JP17/USP) |
| PHENOL [USP-RS] |
| PHENOL [WHO-DD] |
| Phenol, detached crystals |
| PHENOL [MI] |
| Phenol, >=99% |
| WLN: QR |
| Liquefied phenol (JP17) |
| bmse000290 |
| bmse010026 |
| C6H5OH |
| Fenol(DUTCH, POLISH) |
| PHENOL, 80% in ethanol |
| Phenol, LR, >=99% |
| Phenol, natural rubber resin |
| Phen-2,4,6-d3-ol-d |
| MLS001065591 |
| Phenol (CGA 73330) |
| Phenol, for molecular biology |
| BIDD:ER0293 |
| Phenol for disinfection (TN) |
| Phenol, natural, 97%, FG |
| phenylic acid, phenyl hydroxide |
| Cuticura pain relieving ointment |
| CARBOLICUM ACIDUM [HPUS] |
| Phenol, AR, >=99.5% |
| PHENOL,LIQUIFIED [VANDF] |
| BDBM26187 |
| CHEBI:33853 |
| Phenol for disinfection (JP17) |
| (C6-H6-O)x- |
| 3f39 |
| Phenol 10 microg/mL in Methanol |
| METHYL SALICYLATE IMPURITY B |
| EINECS 262-972-4 |
| EINECS 266-017-2 |
| Phenol, Glass Distilled Under Argon |
| Tox21_113463 |
| Tox21_201639 |
| Tox21_300042 |
| LS-476 |
| NA1671 |
| NA2312 |
| NA2821 |
| Phenol 5000 microg/mL in Methanol |
| STL194294 |
| Phenol, solid [UN1671] [Poison] |
| AKOS000119025 |
| Phenol, molten [UN2312] [Poison] |
| Tox21_113463_1 |
| CS-T-39288 |
| DB03255 |
| NA 2821 |
| Phenol, BioXtra, >=99.5% (GC) |
| Phenol, SAJ first grade, >=98.0% |
| UN 1671 |
| UN 2312 |
| UN 2821 |
| USEPA/OPP Pesticide Code: 064001 |
| NCGC00091454-01 |
| NCGC00091454-02 |
| NCGC00091454-03 |
| NCGC00091454-05 |
| NCGC00091454-06 |
| NCGC00091454-07 |
| NCGC00254019-01 |
| NCGC00259188-01 |
| Phenol, JIS special grade, >=99.0% |
| 68071-29-4 |
| AM802906 |
| BP-30160 |
| METHYL SALICYLATE IMPURITY B [EP] |
| SMR000568492 |
| Phenol 1000 microg/mL in Dichloromethane |
| Phenol, PESTANAL(R), analytical standard |
| Liquified Phenol (contains 7-10 % water) |
| LS-105199 |
| FT-0645154 |
| FT-0673707 |
| FT-0693833 |
| P1610 |
| P2771 |
| EN300-19432 |
| C00146 |
| D00033 |
| EC 266-017-2 |
| Estratti, Olio di Catrame di Carbone, Alcalini |
| Phenol, unstabilized, ReagentPlus(R), >=99% |
| Phenol, p.a., ACS reagent, 99.5-100.5% |
| Q130336 |
| J-610001 |
| Phenol, for molecular biology, ~90% (T), liquid |
| A13-01814 |
| F1908-0106 |
| Phenol, unstabilized, purified by redistillation, >=99% |
| Z104473830 |
| InChI=1/C6H6O/c7-6-4-2-1-3-5-6/h1-5,7 |
| Phenol, BioUltra, for molecular biology, >=99.5% (GC) |
| Phenol, United States Pharmacopeia (USP) Reference Standard |
| Liquified Phenol, meets USP testing specifications, >=89.0% |
| Phenol, BioUltra, for molecular biology, TE-saturated, ~73% (T) |
| phenol;phenol [jan];phenol, pure;phenol phenol [jan] phenol, pure |
| Phenol, puriss. p.a., ACS reagent, reag. Ph. Eur., 99.0-100.5% |
| Phenol, contains hypophosphorous as stabilizer, loose crystals, ACS reagent, >=99.0% |
| Phenol, puriss., meets analytical specification of Ph. Eur., BP, USP, 99.5-100.5% (GC) |
| Phenol, puriss., meets analytical specification of Ph. Eur., BP, USP, >=99.5% (GC), crystalline (detached) |
|
There are more than 10 synonyms. If you wish to see them all click here.
|