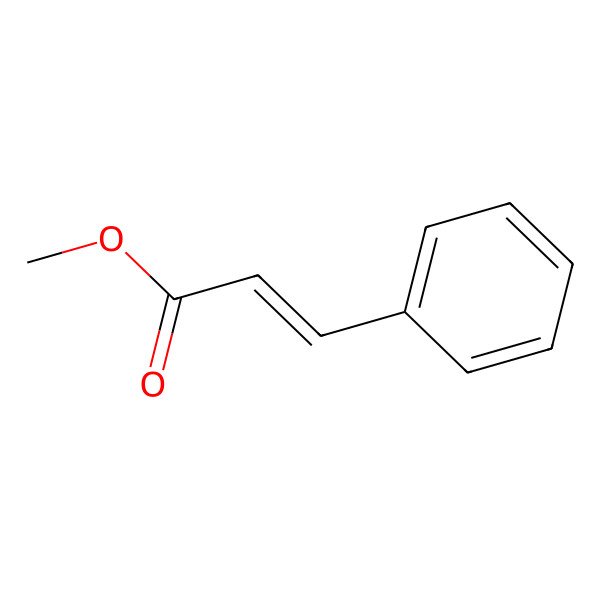
| 103-26-4 |
| Methyl trans-cinnamate |
| 1754-62-7 |
| Methyl (E)-cinnamate |
| Methyl (E)-3-phenylprop-2-enoate |
| Cinnamic acid methyl ester |
| Methyl 3-phenylpropenoate |
| Methyl cinnamylate |
| Methyl 3-phenylacrylate |
| Methylcinnamate |
| trans-Methyl cinnamate |
| (E)-Methyl cinnamate |
| Methyl trans-3-phenyl-2-propenoate |
| methyl-3-phenylprop-2-enoate |
| Methyl 3-phenyl-2-propenoate |
| FEMA No. 2698 |
| methyl (2E)-3-phenylprop-2-enoate |
| trans-Cinnamic acid methyl ester |
| Methyl cinnamate (natural) |
| CINNAMIC ACID, METHYL ESTER |
| 2-Propenoic acid, 3-phenyl-, methyl ester |
| SemaSORB 9815 |
| NSC 9411 |
| Methyl (E)-3-phenyl-2-propenoate |
| 2-PROPENOIC ACID, 3-PHENYL-, METHYL ESTER, (E)- |
| trans-Methyl 3-Phenyl-2-propenoate |
| Cinnamic acid, methyl ester, (E)- |
| EINECS 203-093-8 |
| UNII-533CV2ZCQL |
| (E)-3-Phenylacrylic acid methyl ester |
| 533CV2ZCQL |
| 2-propenoic acid, 3-phenyl-, methyl ester, (2E)- |
| methyl 3-phenylprop-2-enoate |
| AI3-00579 |
| CHEBI:6857 |
| Methyl (2E)-3-phenylacrylate |
| Methyl (E)-3-phenylpropenoate |
| DTXSID5044314 |
| NSC-9411 |
| EC 203-093-8 |
| cis-methyl cinnamate |
| Methyl cinnamate, (E) |
| DTXSID6042151 |
| Methyl (2E)-3-phenyl-2-propenoate |
| 3-Phenyl-2-propenoic acid methyl ester |
| Cinnamic acid methyl |
| Methyl(E)-cinnamate |
| MFCD00008458 |
| methyl trans cinnamate |
| methyl-trans-cinnamate |
| Nat. Methyl Cinnamate |
| WLN: 1OV1U1R |
| ghl.PD_Mitscher_leg0.369 |
| methyl-3 phenylprop-2-enoate |
| CHEMBL55060 |
| Methyl trans-3-Phenylacrylate |
| Methyl trans-cinnamate, 99% |
| SCHEMBL101530 |
| METHYL CINNAMATE [FCC] |
| (e)-cinnamic acid methyl ester |
| Benzeneacrylic acid methyl ester |
| METHYL CINNAMATE [FHFI] |
| DTXCID4022151 |
| DTXCID5028118 |
| METHYL CINNAMATE, (E)- |
| Methyl (2E)-3-phenylpropenoate |
| NSC9411 |
| 3-Phenyl-2-propenoic acid methyl |
| CHEBI:194138 |
| Tox21_301384 |
| Tox21_304025 |
| s6221 |
| STL582416 |
| Methyl cinnamate, >=99.0% (GC) |
| AKOS015890136 |
| CS-W017928 |
| HY-W017212 |
| LS-2916 |
| CINNAMIC ACID METHYL ESTER [MI] |
| Methyl (2E)-3-phenyl-2-propenoate # |
| trans-3-Phenylacrylic Acid Methyl Ester |
| NCGC00255910-01 |
| NCGC00357235-01 |
| CAS-103-26-4 |
| DS-17838 |
| CAS-1754-62-7 |
| Methyl trans-cinnamate, analytical standard |
| Methyl trans-cinnamate, >=98%, FCC, FG |
| (E)-3-Phenyl-2-propenoic acid methyl ester |
| (E)-methyl ester 3-phenyl-2-propenoic acid |
| (2E)-3-Phenyl-2-propenoic acid methyl ester |
| EN300-91678 |
| trans-3-Phenyl-2-propenoic acid methyl ester |
| C06358 |
| EN300-365971 |
| Methyl cinnamate, analytical reference material |
| Methyl cinnamate, natural, >=98%, FCC, FG |
| P16737 |
| trans-3-phenyl-prop-2-enoic acid methyl ester |
| Q204178 |
| J-000917 |
| J-011115 |
| J-522598 |
| Q-100258 |
| Z19782655 |
| InChI=1/C10H10O2/c1-12-10(11)8-7-9-5-3-2-4-6-9/h2-8H,1H3/b8-7 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|