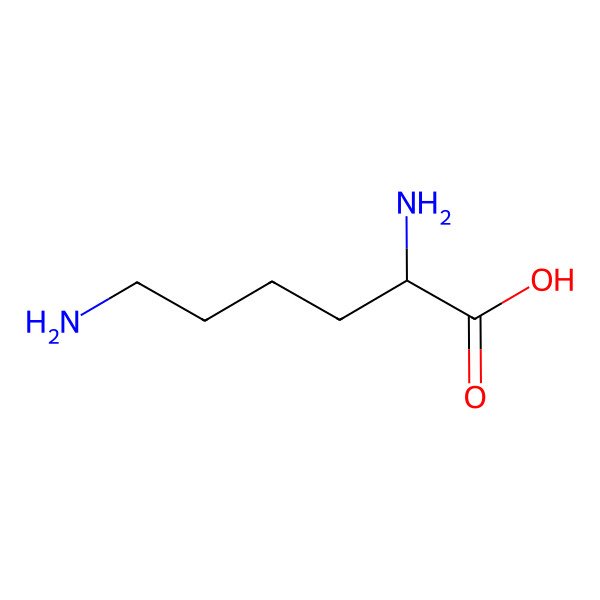
| lysine |
| 56-87-1 |
| lysine acid |
| h-Lys-oh |
| (S)-Lysine |
| (2S)-2,6-diaminohexanoic acid |
| Aminutrin |
| alpha-Lysine |
| L-(+)-Lysine |
| (S)-2,6-Diaminohexanoic acid |
| Hydrolysin |
| Lysinum |
| (S)-alpha,epsilon-Diaminocaproic acid |
| Lysinum [Latin] |
| L-lys |
| Lisina [Spanish] |
| L-Norleucine, 6-amino- |
| lysin |
| Lysine, L- |
| Lisina |
| LYS (IUPAC abbreviation) |
| Lysine [USAN:INN] |
| (S)-2,6-Diaminocaproic acid |
| 25104-18-1 |
| lys |
| CHEBI:18019 |
| Hexanoic acid, 2,6-diamino-, (S)- |
| L-2,6-Diaminocaproic acid |
| 2,6-Diaminohexanoic acid, (S)- |
| lysina |
| (+)-S-Lysine |
| BRN 1722531 |
| HSDB 2108 |
| a-Lysine |
| AI3-26523 |
| L-Lysin |
| EINECS 200-294-2 |
| 3H-Lysine |
| UNII-K3Z4F929H6 |
| DTXSID6023232 |
| 6-ammonio-L-norleucine |
| K3Z4F929H6 |
| 12798-06-0 |
| DTXCID403232 |
| L-Lysine, labeled with tritium |
| L-Lysine base |
| Lysinum (Latin) |
| 4-04-00-02717 (Beilstein Handbook Reference) |
| LYSINE (II) |
| LYSINE [II] |
| LYSINE (MART.) |
| LYSINE [MART.] |
| L-2,6-Diaminocaproate |
| L-LYSINE, MONOACETATE |
| MFCD00064433 |
| 20166-34-1 |
| TYROSINE IMPURITY B (EP IMPURITY) |
| TYROSINE IMPURITY B [EP IMPURITY] |
| 2,6-diaminohexanoate |
| alpha,epsilon-Diaminocaproic acid |
| Gidrolizin |
| Hydrolysine |
| (C6-H14-N2-O2)x- |
| l- lysine |
| L-Lisina |
| .alpha.-Lysine |
| 1ozv |
| 1yxd |
| 3h-l-lysine |
| 6-amino-Aminutrin |
| NCGC00164527-01 |
| L-?Lysine |
| H-Lys |
| (-)-lysine |
| 6-amino-L-Norleucine |
| Lysine, L-, peptides |
| a,e-Diaminocaproic acid |
| Lysine (USAN/INN) |
| L-2,6-Diainohexanoate |
| LYSINE [VANDF] |
| LYSINE [HSDB] |
| LYSINE [INCI] |
| LYSINE [USAN] |
| LYSINE [INN] |
| L-LYSINE [FHFI] |
| LYSINE [WHO-DD] |
| (S)-a,e-Diaminocaproate |
| LYSINE [MI] |
| L-Lysine, >=97% |
| bmse000043 |
| bmse000914 |
| D07TMZ |
| Epitope ID:136017 |
| 2,6-Diamino hexanoic acid |
| (S)-2,6-Diaminohexanoate |
| L-2,6-Diainohexanoic acid |
| CHEMBL8085 |
| GTPL724 |
| (S)-2,6-diamino-Hexanoate |
| (S)-a,e-Diaminocaproic acid |
| L-Lysine, analytical standard |
| L-Lysine, >=98%, FG |
| (S)-2,6-diamino-Hexanoic acid |
| B05XB03 |
| L-Lysine, >=98% (TLC) |
| alpha, epsilon-Diaminocaproic acid |
| BDBM217367 |
| (2S)-2,6-Diamino-hexanoic acid |
| HY-N0469 |
| L-H2N(CH2)4CH(NH2)COOH |
| Tox21_112158 |
| Ethyl3,5-dichloro-4-propoxybenzoate |
| s5630 |
| .alpha.,.epsilon.-Diaminocaproic acid |
| AKOS006239081 |
| AKOS015855172 |
| CCG-266180 |
| CS-W019758 |
| DB00123 |
| CAS-56-87-1 |
| NCGC00166296-02 |
| 26714-32-9 |
| AC-14492 |
| AS-11733 |
| LS-88418 |
| (S)-.alpha.,.epsilon.-Diaminocaproic acid |
| L-Lysine, crystallized, >=98.0% (NT) |
| AM20100376 |
| L-103 |
| L0129 |
| L-Lysine, Vetec(TM) reagent grade, >=98% |
| A20652 |
| C00047 |
| D02304 |
| A904498 |
| A919375 |
| J-521651 |
| (S)-2,6-Diaminocaproic acid;(S)-(+)-Lysine;Lysine |
| Q20816880 |
| F0001-1472 |
| 0013CD6B-1671-4369-B1BE-F531611E50C7 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|