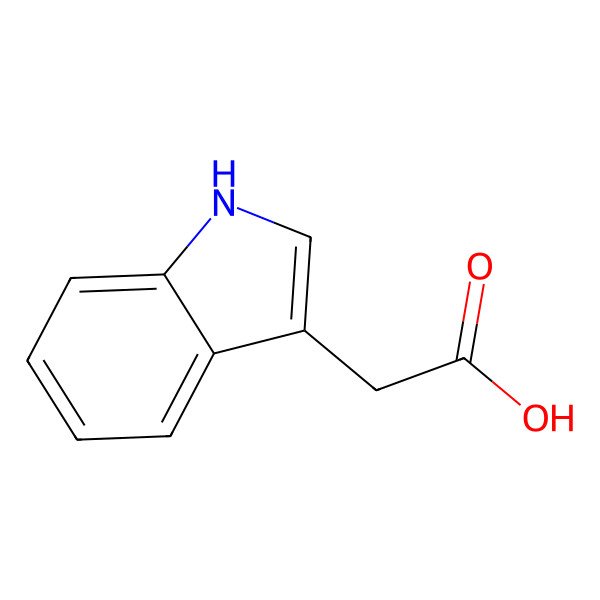
| 87-51-4 |
| 3-Indoleacetic acid |
| indoleacetic acid |
| Heteroauxin |
| 1H-Indole-3-acetic acid |
| 1H-indol-3-ylacetic acid |
| 2-(1H-Indol-3-yl)acetic acid |
| Rhizopin |
| Rhizopon A |
| Indol-3-ylacetic acid |
| 3-Iaa |
| 3-(Carboxymethyl)indole |
| 3-Indolylacetic acid |
| auxin |
| Hexteroauxin |
| beta-Indoleacetic acid |
| indoleacetate |
| Acetic acid, indolyl- |
| Heteroauxinhexteroauxiniaa |
| IAA |
| Indolylacetic acid |
| beta-Indolylacetic acid |
| Indolyl-3-acetic acid |
| indole-3-acetate |
| indole acetic acid |
| omega-Skatole carboxylic acid |
| Kyselina 3-indolyloctova |
| (1H-Indol-3-yl)-acetic acid |
| Indoleacetic acid (VAN) |
| (indol-3-yl)acetate |
| CCRIS 1014 |
| EPA Pesticide Chemical Code 128915 |
| Kyselina 3-indolyloctova [Czech] |
| (indol-3-yl)acetic acid |
| AI3-24131 |
| 2-(3-Indolyl)acetic acid |
| NSC 3787 |
| 2-(indol-3-yl)ethanoic acid |
| 3-Indolylessigsaeure |
| .alpha.-IAA |
| .beta.-IAA |
| 3-indole acetic acid |
| Indole 3-acetic acid |
| 3-Indole-Acetic acid |
| .beta.-Indoleacetic acid |
| EINECS 201-748-2 |
| MFCD00005636 |
| .beta.-Indolylacetic acid |
| .beta.-Indole-3-acetic acid |
| CHEMBL82411 |
| (1H-Indol-3-yl)acetic acid |
| UNII-6U1S09C61L |
| .omega.-Skatole carboxylic acid |
| DTXSID5020738 |
| CHEBI:16411 |
| .alpha.-Indol-3-yl-acetic acid |
| Indole--d5-3-acetic--d2 Acid |
| NSC3787 |
| 1H-Indole-3-acetic acid (9CI) |
| 6U1S09C61L |
| NSC-3787 |
| 3-indoleacetate |
| 1H-Indole-3-acetate |
| 1173020-21-7 |
| DTXCID70738 |
| (1H-indol-3-yl)acetate |
| CAS-87-51-4 |
| IES |
| SMR000471855 |
| Indolylacetate |
| b-Indoleacetate |
| b-Indolylacetate |
| 3-Indolylacetate |
| alpha-IAA |
| beta-Indoleacetate |
| beta-IAA |
| beta-Indolylacetate |
| Indol-3-ylacetate |
| Indolyl-3-acetate |
| Skatole carboxylate |
| b-Indoleacetic acid |
| IAC |
| b-Indolylacetic acid |
| Indole-3acetic acid |
| indol-3-yleddikesyre |
| indole-3-acetic aicd |
| 3-indolyl acetic acid |
| 3-indolyl-acetic acid |
| Skatole carboxylic acid |
| Acid, 6 |
| 2-(3-Indolyl)acetate |
| Indole-3-acetic-t acid |
| 1H-indol-3-acetic acid |
| Maybridge1_006755 |
| 1H-Indole 3-acetic acid |
| beta-Indole-3-acetic acid |
| bmse000177 |
| (1H-Indol-3-yl)-acetate |
| 3-Indoleacetic acid, 98% |
| 3-Indoleacetic acid, 99% |
| Oprea1_602123 |
| SCHEMBL26344 |
| MLS001066408 |
| MLS001331664 |
| MLS001332399 |
| MLS001332400 |
| 3-Indolylmethylcarboxylic acid |
| alpha-Indol-3-yl-acetic acid |
| Indole-3-acetic acid (8CI) |
| INDOLEACETIC ACID [MI] |
| WLN: T56 BMJ D1VQ |
| [3H]-IAA |
| HMS560L01 |
| AMY2736 |
| INDOLE ACETIC ACID [INCI] |
| 2-(1H-indol-3-yl)-acetic acid |
| HMS2269G24 |
| HMS3604M04 |
| BCP26623 |
| Tox21_202284 |
| Tox21_302731 |
| BBL009348 |
| BDBM50201883 |
| CCG-51070 |
| s4799 |
| STK397461 |
| AKOS000119890 |
| 1H-Indole-3-acetic-a-t acid (9CI) |
| AC-2974 |
| CG-0522 |
| CS-6287 |
| DB07950 |
| LS-7456 |
| SDCCGMLS-0066204.P001 |
| 3-Indoleacetic acid, >=98.0% (T) |
| NCGC00247039-01 |
| NCGC00247039-02 |
| NCGC00256391-01 |
| NCGC00259833-01 |
| HY-18569 |
| SY003464 |
| BB 0242380 |
| EU-0099905 |
| FT-0627215 |
| FT-0695803 |
| I0022 |
| 3-Indoleacetic acid, technical, >=95% (T) |
| EN300-17303 |
| C00954 |
| I-1000 |
| I-1040 |
| Q411208 |
| SR-01000596909 |
| 2-(1H-indol-3-yl)acetic acid;Indole-3-acetic acid |
| 3-Indoleacetic acid, SAJ special grade, >=98.5% |
| IS_INDOLE-2,4,5,6,7-D5-3-ACETIC ACID |
| SR-01000596909-1 |
| SR-01000596909-2 |
| 3-Indolyl acetic acid 100 microg/mL in Acetonitrile |
| Z56913182 |
| 2-(3-Indolyl)acetic acid 3-(Carboxymethyl)-1H-indole |
| 3-Indoleacetic acid, PESTANAL(R), analytical standard |
| F0722-8837 |
| 0A0524AE-D755-4E91-A73A-2AD867FE676A |
| 3-Indoleacetic acid, plant cell culture tested, crystalline |
| InChI=1/C10H9NO2/c12-10(13)5-7-6-11-9-4-2-1-3-8(7)9/h1-4,6,11H,5H2,(H,12,13 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|