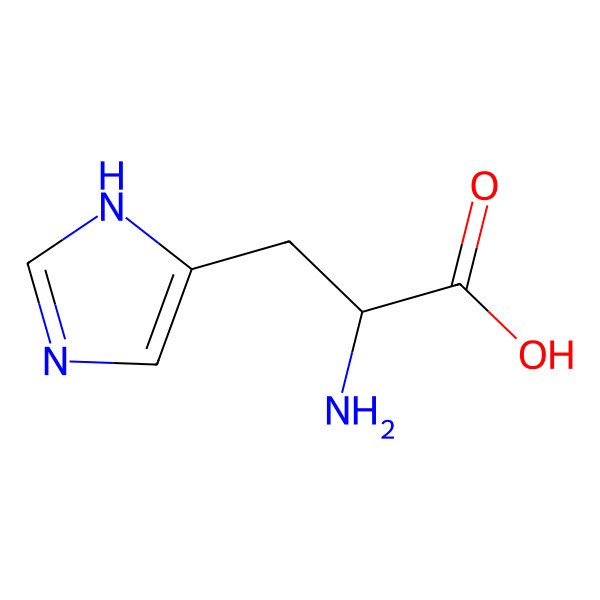
| histidine |
| 71-00-1 |
| H-His-OH |
| glyoxaline-5-alanine |
| L-(-)-Histidine |
| Anti-rheuma |
| Istidina |
| S-Histidine |
| (L)-Histidine |
| histidina |
| Histidinum |
| HISTIDINE, L- |
| L-Histidin |
| (S)-4-(2-Amino-2-carboxyethyl)imidazole |
| Histidine (VAN) |
| (2S)-2-amino-3-(1H-imidazol-4-yl)propanoic acid |
| FEMA No. 3694 |
| L-beta-(4-Imidazolyl)alanin |
| (S)-alpha-amino-1H-imidazole-4-propanoic acid |
| Histidine [USAN:INN] |
| Histidinum [INN-Latin] |
| 4-(2-Amino-2-carboxyethyl)imidazole |
| (S)-Histidine |
| his |
| Histidina [INN-Spanish] |
| 1H-Imidazole-4-alanine, (S)- |
| L-His |
| (S)-2-Amino-3-(4-imidazolyl)propionsaeure |
| L-Alanine, 3-(1H-imidazol-4-yl)- |
| L-hystidine |
| HSDB 1810 |
| L-Hisidine |
| AI3-26558 |
| 7006-35-1 |
| EINECS 200-745-3 |
| L-beta-(4-Imidazolyl)-alpha-alanin |
| (S)-alpha-Amino-1H-imidazole-4-propionic acid |
| NSC 137773 |
| UNII-4QD397987E |
| CHEBI:15971 |
| alpha-Amino-4(or 5)-imidazolepropionic acid |
| Histidine (L-Histidine) |
| MFCD00064315 |
| 4QD397987E |
| NSC-137773 |
| alpha-Amino-1H-imidazole-4-propionic acid, (S)- |
| 1H-Imidazole-4-propanoic acid, alpha-amino-, (S)- |
| (S)-a-Amino-1H-imidazole-4-propanoic acid |
| DTXSID9023126 |
| (S)-2-Amino-3-(4-imidazolyl)propionic acid |
| (2S)-2-amino-3-(imidazol-4-yl)propanoic acid |
| [3H]histidine |
| HISTIDINE (II) |
| HISTIDINE [II] |
| [3H]-histidine |
| HISTIDINE (MART.) |
| HISTIDINE [MART.] |
| histidin |
| (S)-2-amino-3-(1H-imidazol-4-yl)propanoic acid |
| HISTIDINE (EP MONOGRAPH) |
| HISTIDINE [EP MONOGRAPH] |
| HISTIDINE (USP MONOGRAPH) |
| HISTIDINE [USP MONOGRAPH] |
| SERINE IMPURITY C (EP IMPURITY) |
| SERINE IMPURITY C [EP IMPURITY] |
| L-isomer Histidine |
| Histidine, L isomer |
| Histidine, L-isomer |
| (2S)-2-amino-3-(1H-imidazol-5-yl)propanoic acid |
| CHEBI:27570 |
| L-histidina |
| NSC137773 |
| 1hsl |
| 1lag |
| L-Histidine Base |
| Histidine,(S) |
| (2S)-2-amino-3-(1H-imidazol-4-yl)propanoic acid hydrochloride |
| Histidine (USP/INN) |
| HISTIDINE [INN] |
| HISTIDINE [MI] |
| L-Histidine (JP17) |
| L-2-Amino-3-(4-imidazolyl)propionic acid |
| HISTIDINE [HSDB] |
| HISTIDINE [INCI] |
| HISTIDINE [USAN] |
| HISTIDINE [VANDF] |
| Lopac-H-8125 |
| bmse000039 |
| bmse000976 |
| bmse001015 |
| D0T6UL |
| L-HISTIDINE [FCC] |
| L-HISTIDINE [JAN] |
| L-Histidine (H-His-OH) |
| amino-4-imidazoleproprionate |
| HISTIDINE [WHO-DD] |
| L-HISTIDINE [FHFI] |
| SCHEMBL3259 |
| Lopac0_000566 |
| US9138393, Histidine |
| US9144538, Histidine |
| (S)1H-Imidazole-4-alanine |
| CHEMBL17962 |
| L-HISTIDINE [USP-RS] |
| (S)-1H-Imidazole-4-alanine |
| BDBM7953 |
| DTXCID503126 |
| GTPL3310 |
| GTPL4670 |
| L-Histidine, non-animal source |
| Imidazole C-4(5) deriv. 5 |
| amino-4-imidazoleproprionic acid |
| amino-1H-imidazole-4-propanoate |
| L-Histidine, p.a., 98.5% |
| BDBM181118 |
| (C6-H9-N3-O2)x- |
| 3-(1H-imidazol-4-yl)-L-Alanine |
| HY-N0832 |
| amino-1H-imidazole-4-propanoic acid |
| AKOS015854051 |
| AKOS026676613 |
| AM81801 |
| CCG-204656 |
| CS-7781 |
| DB00117 |
| LS-2343 |
| SDCCGSBI-0050549.P002 |
| (S)-a-Amino-1H-imidazole-4-propanoate |
| NCGC00015518-01 |
| NCGC00162189-01 |
| NCGC00162189-02 |
| NCGC00162189-05 |
| AC-35086 |
| AS-14171 |
| (S)-alpha-Amino-1H-imidazole-4-propanoate |
| (S)-alpha-Amino-1H-imidazole-4-propionate |
| L-Histidine, BioUltra, >=99.5% (NT) |
| H0149 |
| L-Histidine, SAJ special grade, >=98.5% |
| S3989 |
| EN300-57334 |
| C00135 |
| D00032 |
| D70843 |
| H-2310 |
| L-Histidine, ReagentPlus(R), >=99% (TLC) |
| M02982 |
| L-Histidine, Vetec(TM) reagent grade, >=99% |
| Q485277 |
| B81AEDB0-EACA-4296-9BAB-52D60F137FFB |
| 1H-Imidazole-4-propanoic acid, .alpha.-amino-, (S)- |
| F8881-8926 |
| F8889-0575 |
| L-Histidine, certified reference material, TraceCERT(R) |
| Z359369984 |
| Histidine, European Pharmacopoeia (EP) Reference Standard |
| L-Histidine, United States Pharmacopeia (USP) Reference Standard |
| L-Histidine, Pharmaceutical Secondary Standard; Certified Reference Material |
| L-Histidine, cell culture tested, meets EP, USP testing specifications, from non-animal source |
|
There are more than 10 synonyms. If you wish to see them all click here.
|