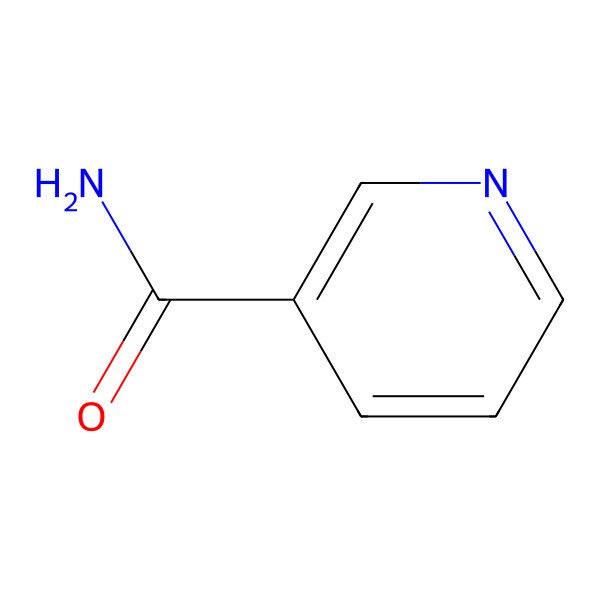
| niacinamide |
| 98-92-0 |
| 3-Pyridinecarboxamide |
| Nicotinic acid amide |
| pyridine-3-carboxamide |
| vitamin PP |
| Papulex |
| Aminicotin |
| Amixicotyn |
| Nicobion |
| Nicotylamide |
| Nikotinamid |
| Savacotyl |
| Benicot |
| Dipegyl |
| Endobion |
| Hansamid |
| Pelmine |
| Nicotinic amide |
| Delonin amide |
| Pelonin amide |
| Vi-Nicotyl |
| Austrovit PP |
| Inovitan PP |
| Nicosylamide |
| Nicotilamide |
| Nicotililamido |
| Amnicotin |
| Niacevit |
| Nicamina |
| Nicamindon |
| Nicofort |
| Nicomidol |
| Nicotamide |
| Nicovitina |
| Nicovitol |
| Nicozymin |
| Niocinamide |
| Niozymin |
| Niamide |
| Nicasir |
| Nicogen |
| Nicota |
| Nicotol |
| Nicovit |
| Niko-tamin |
| 3-Carbamoylpyridine |
| Nicotine acid amide |
| Nandervit-N |
| Pyridine-3-carboxylic acid amide |
| Vitamin B |
| Niavit PP |
| Nicotinamidum |
| Nicosan 2 |
| Nicotine amide |
| beta-Pyridinecarboxamide |
| Nikotinsaeureamid |
| Nicotylamidum |
| Mediatric |
| Nicotinsaureamid |
| Pyridine, 3-carbamoyl- |
| 3-Pyridinecarboxylic acid amide |
| m-(Aminocarbonyl)pyridine |
| Acid amide |
| Factor pp |
| Nicotinamida |
| Nicovel |
| Vitamin B (VAN) |
| Pelmin |
| Amid kyseliny nikotinove |
| Witamina PP |
| PP-Faktor |
| Amide PP |
| Nicotinsaureamid [German] |
| Nikotinsaeureamid [German] |
| Amid kyseliny nikotinove [Czech] |
| Nicotinamidum [INN-Latin] |
| Nicotinamida [INN-Spanish] |
| NAM |
| Niacinamide [USAN] |
| C6H6N2O |
| Niacinamid |
| Nictoamide |
| CCRIS 1901 |
| Dipigyl |
| HSDB 1237 |
| Vi-noctyl |
| 3-(aminocarbonyl)pyridine |
| AI3-02906 |
| NSC 13128 |
| b-Pyridinecarboxamide |
| Niacinamide [USP] |
| Nicotinamide [INN] |
| EINECS 202-713-4 |
| MFCD00006395 |
| NSC-13128 |
| NSC-27452 |
| .beta.-Pyridinecarboxamide |
| Nicotinamide-(amide-15N) |
| Nicotinamide (Vitamin B3) |
| CHEMBL1140 |
| UNII-25X51I8RD4 |
| MLS000069714 |
| DEA No. 1405 |
| DTXSID2020929 |
| CHEBI:17154 |
| 25X51I8RD4 |
| NSC13128 |
| Niacinamide (USP) |
| NCGC00093354-03 |
| NCGC00093354-05 |
| SMR000058212 |
| Nicotinamide 10 microg/mL in Acetonitrile |
| Niacinamide;Nicotinic acid amide;Vitamin B3 |
| WLN: T6NJ CVZ |
| EC 202-713-4 |
| DTXCID00929 |
| CAS-98-92-0 |
| SR-01000721872 |
| nicotin-amide |
| 3-Amidopyridine |
| Nicotinamide,(S) |
| Vitamin B3 amide |
| 3-yridinecarboxamide |
| 3-piridinacarboxamida |
| Mediatric (Salt/Mix) |
| niacin - Vitamin B3 |
| 1yc5 |
| Opera_ID_775 |
| Niacin (as niacinamide) |
| NIACINAMIDE [II] |
| Niacinamide(Vitamin B3) |
| NIACINAMIDE [FCC] |
| NICOTINAMIDE [MI] |
| Niacinamide (nicotinamide) |
| NIACINAMIDE [HSDB] |
| NIACINAMIDE [INCI] |
| NICOTINAMIDE [JAN] |
| bmse000281 |
| MolMap_000061 |
| NIACINAMIDE [VANDF] |
| SCHEMBL2926 |
| Nicotinamide (JP17/INN) |
| NICOTINAMIDUM [HPUS] |
| NIACINAMIDE [USP-RS] |
| NICOTINAMIDE [MART.] |
| MLS001424246 |
| NICOTINAMIDE [WHO-DD] |
| NICOTINAMIDE [WHO-IP] |
| SCHEMBL6278767 |
| SGCUT00176 |
| TPN COMPONENT NIACINAMIDE |
| SCHEMBL19978192 |
| BDBM27507 |
| NIACINAMIDE [ORANGE BOOK] |
| HMS2052M21 |
| HMS2090B05 |
| HMS2093H03 |
| HMS2236J03 |
| HMS3370F21 |
| HMS3394M21 |
| HMS3655M20 |
| HMS3713B22 |
| HMS3884A16 |
| NICOTINAMIDE [EP IMPURITY] |
| Pharmakon1600-01505397 |
| NIACINAMIDE [USP MONOGRAPH] |
| NICOTINAMIDE [EP MONOGRAPH] |
| BCP07322 |
| HY-B0150 |
| NIACINAMIDE COMPONENT OF TPN |
| NSC27452 |
| to_000073 |
| EINECS 234-265-0 |
| Nicotinamide 1.0 mg/ml in Methanol |
| Nicotinamide, >=98.5% (HPLC) |
| Nicotinamide, >=99.5% (HPLC) |
| Tox21_111202 |
| Tox21_201716 |
| Tox21_302776 |
| NICOTINAMIDUM [WHO-IP LATIN] |
| NSC759115 |
| s1899 |
| STL163867 |
| AKOS005715850 |
| Tox21_111202_1 |
| CCG-101149 |
| CS-1968 |
| CS-O-30679 |
| DB02701 |
| LS-2051 |
| NC00399 |
| NSC-759115 |
| SB74497 |
| Nicotinamide 100 microg/mL in Methanol |
| Nicotinamide, >=98% (HPLC), powder |
| NCGC00093354-04 |
| NCGC00093354-06 |
| NCGC00093354-09 |
| NCGC00256432-01 |
| NCGC00259265-01 |
| NIACIN (AS NIACINAMIDE) [VANDF] |
| 11032-50-1 |
| AS-13845 |
| BN166252 |
| Nicotinamide, puriss., 99.0-101.0% |
| SY024804 |
| Nicotinamide, tested according to Ph.Eur. |
| SBI-0206826.P001 |
| FT-0631517 |
| FT-0672696 |
| FT-0773644 |
| N0078 |
| SW197779-3 |
| EN300-15612 |
| Niacinamide, meets USP testing specifications |
| C00153 |
| D00036 |
| Nicotinamide (Niacinamide), analytical standard |
| AB00373895-13 |
| AB00373895_15 |
| AB00373895_16 |
| Nicotinamide, Vetec(TM) reagent grade, >=98% |
| A845925 |
| AC-907/25014114 |
| Q192423 |
| Q-201470 |
| SR-01000721872-3 |
| SR-01000721872-4 |
| SR-01000721872-5 |
| Z33546463 |
| F2173-0513 |
| Niacinamide;Nicotinic acid amide;Vitamin B3; Vitamin PP |
| Nicotinamide, British Pharmacopoeia (BP) Reference Standard |
| A186B02E-6C70-4E54-9739-79398D439AAA |
| Nicotinamide, European Pharmacopoeia (EP) Reference Standard |
| Niacinamide, United States Pharmacopeia (USP) Reference Standard |
| InChI=1/C6H6N2O/c7-6(9)5-2-1-3-8-4-5/h1-4H,(H2,7,9 |
| Niacinamide, Pharmaceutical Secondary Standard; Certified Reference Material |
| Nicotinamide, BioReagent, suitable for cell culture, suitable for insect cell culture |
| 63748-44-7 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|