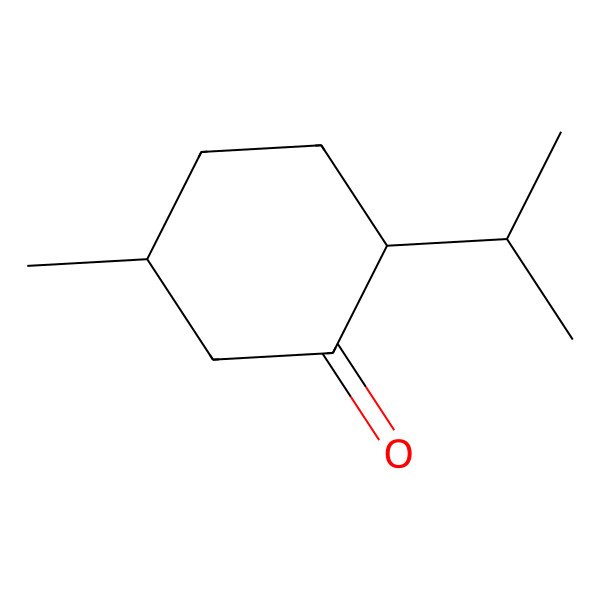
| MENTHONE |
| 14073-97-3 |
| l-MENTHONE |
| (2S,5R)-2-isopropyl-5-methylcyclohexanone |
| trans-Menthone |
| p-Menthone |
| Neomenthone |
| Menthone racemic |
| L-Menthan-3-one |
| trans-Menthan-3-one |
| 89-80-5 |
| trans-p-Menthan-3-one |
| (1R,4S)-(-)-p-Menthan-3-one |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, (2S,5R)- |
| DL-Menthone |
| Menthone (natural) |
| p-Menthan-3-one, trans- |
| FEMA No. 2667 |
| p-Menthan-3-one racemic |
| (dl)-Menthone |
| (-)-(2S,5R)-Menthone |
| (1R,4S)-p-menthan-3-one |
| CCRIS 5747 |
| (2S,5R)-5-methyl-2-(propan-2-yl)cyclohexanone |
| HSDB 1268 |
| p-Menthan-3-one, dl- |
| EINECS 201-941-1 |
| EINECS 214-049-2 |
| UNII-9NH5J4V8FN |
| (2S,5R)-5-methyl-2-(1-methylethyl)cyclohexanone |
| (2S,5R)-5-methyl-2-propan-2-ylcyclohexan-1-one |
| (2S-trans)-5-methyl-2-(1-methylethyl)cyclohexanone |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, trans- |
| (-)-(1R,4S)-menthone |
| UNII-5F709W4OG4 |
| AI3-11106 |
| (2S,5R)-5-METHYL-2-(1-METHYLETHYL)-CYCLOHEXANONE |
| DTXSID3044384 |
| CHEBI:15410 |
| (-)-5-Methyl-2-(1-methylethyl)cyclohexanone |
| 5-Methyl-2-(1-methylethyl)cyclohexanone, trans- |
| 5F709W4OG4 |
| Cyclohexan-1-one, 2-isopropyl-5-methyl-, racemic |
| EINECS 237-926-1 |
| (2S,5R)-5-Methyl-2-(Propan-2-Yl)Cyclohexan-1-One |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, (2S-trans)- |
| EC 237-926-1 |
| trans-menthon |
| DTXCID1024384 |
| trans - menthone |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, trans-(+/-)- |
| l-p-Menthan-3-one |
| 5-Methyl-2-(1-methylethyl)cyclohexanone |
| CAS-14073-97-3 |
| menthon |
| Menthone G |
| trans-p-menthone |
| 1-menthone |
| (-)menthone |
| NCGC00095606-01 |
| MFCD00001634 |
| Menthan-3-one, trans |
| Spectrum_001299 |
| rel-(2R,5S)-2-isopropyl-5-methylcyclohexanone |
| (-)-Menthone,(S) |
| MENTHONE [FHFI] |
| SpecPlus_000437 |
| MENTHONE, L- |
| L-MENTHONE [MI] |
| Spectrum2_000691 |
| Spectrum3_001272 |
| Spectrum4_001648 |
| Spectrum5_000495 |
| (2S,5R)-2-isopropyl-5-methyl-cyclohexanone |
| bmse000375 |
| (-)-Menthone, 90% |
| 9NH5J4V8FN |
| SCHEMBL21709 |
| BSPBio_002864 |
| KBioGR_002115 |
| KBioSS_001779 |
| L - menthan - 3 - one |
| MENTHONE, (-)- |
| SPECTRUM300564 |
| DivK1c_006533 |
| SPBio_000841 |
| (1S)-trans-p-menthan-3-one |
| (-)-MENTHONE [FCC] |
| CHEMBL276311 |
| DTXSID2044478 |
| KBio1_001477 |
| KBio2_001779 |
| KBio2_004347 |
| KBio2_006915 |
| KBio3_002364 |
| (-)-Menthone, analytical standard |
| HY-N7916 |
| Tox21_111510 |
| Tox21_302153 |
| CCG-38562 |
| MFCD00136033 |
| AKOS006343213 |
| Tox21_111510_1 |
| LMPR0102090004 |
| LS-2354 |
| SDCCGMLS-0066582.P001 |
| 2-Isopropyl-5-methylcyclohexanone, trans |
| NCGC00095606-02 |
| NCGC00178425-01 |
| NCGC00255957-01 |
| p-Menthan-3-one, (1R,4S)-(-)- |
| AS-17440 |
| (2S,5R)2-isopropyl-5-methylcyclohexanone |
| CS-0138798 |
| 1-METHYL-4-ISOPROPYLCYCLOHEXAN-3-ONE |
| C00843 |
| EN300-383879 |
| (2S,5R)-2-Isopropyl-5-methylcyclohexan-1-one |
| (2S, 5R)-trans-2-isopropyl-5-methylcyclohexanone |
| Q424902 |
| SR-05000002387 |
| 5-Methyl-2-(1-methylethyl)-(2S,5R)-Cyclohexanone |
| SR-05000002387-1 |
| W-108194 |
| 5-Methyl-2-(1-methylethyl)-(2S-trans)-Cyclohexanone |
| (-)-Menthone, primary pharmaceutical reference standard |
| Z1201619246 |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, trans-(.+/-.)- |
| 1/C10H18O/c1-7(2)9-5-4-8(3)6-10(9)11/h7-9H,4-6H2,1-3H3/t8-,9+/m1/s |
| InChI=1/C10H18O/c1-7(2)9-5-4-8(3)6-10(9)11/h7-9H,4-6H2,1-3H3/t8-,9+/m1/s |
|
There are more than 10 synonyms. If you wish to see them all click here.
|