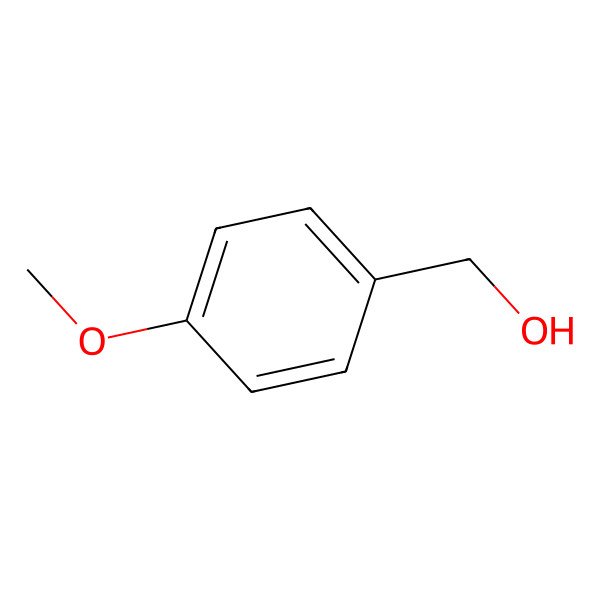
| 105-13-5 |
| (4-Methoxyphenyl)methanol |
| Anisyl alcohol |
| Anise alcohol |
| p-Methoxybenzyl alcohol |
| p-Anisyl alcohol |
| Benzenemethanol, 4-methoxy- |
| Anisic alcohol |
| p-Anisol alcohol |
| 4-Methoxybenzenemethanol |
| Benzyl alcohol, p-methoxy- |
| Anis alcohol |
| Anisalcohol, p- |
| FEMA No. 2099 |
| 4-Anisylalcohol |
| Anisyl alcohol (natural) |
| NSC 2151 |
| CCRIS 5111 |
| p-methoxy-benzyl alcohol |
| EINECS 203-273-6 |
| MFCD00004653 |
| 4-Methoxybenzyl-d2Alcohol |
| (4-methoxyphenyl)-methanol |
| BRN 0636654 |
| Benzenemethanol, ar-methoxy- |
| UNII-7N6XGV3U49 |
| (4-Methoxy-phenyl)-methanol |
| AI3-01170 |
| 7N6XGV3U49 |
| DTXSID6044357 |
| NSC-2151 |
| EC 203-273-6 |
| 35693-15-3 |
| 4-06-00-05909 (Beilstein Handbook Reference) |
| [4-(methyloxy)phenyl]methanol |
| (4-(METHYLOXY)PHENYL)METHANOL |
| METHOXYBENZYLALCOHOL |
| Anisalkohol |
| p-anisalcohol |
| para-anisyl alcohol |
| JandaJel(TM)-Wang |
| p-methoxybenzylalcohol |
| 4-methoxybenzylalkohol |
| 4-methoxyphenylmethanol |
| 4-methoxy-benzylalcohol |
| 4-methoxylbenzyl alcohol |
| 4-methyoxybenzyl alcohol |
| 4-methoxy-benzenemethanol |
| 4-methoxy-benzyl alcohol |
| 4-(Hydroxymethyl)anisole |
| para-methoxybenzyl alcohol |
| 4 - methoxybenzyl alcohol |
| bmse010025 |
| Methoxybenzyl alcohol, 4- |
| ANISE ALCOHOL [MI] |
| Benzyl alcohol, ar-methoxy- |
| DSSTox_CID_24357 |
| DSSTox_RID_82376 |
| 4-methoxyphenylmethyl alcohol |
| DSSTox_GSID_47486 |
| SCHEMBL27329 |
| WLN: Q1R DO1 |
| (4-Methoxyphenyl)methanol # |
| ANISE ALCOHOL [INCI] |
| ANISYL ALCOHOL [FCC] |
| ANISYL ALCOHOL [FHFI] |
| 4-Methoxybenzyl alcohol, 98% |
| CHEMBL294431 |
| DTXCID4024357 |
| FEMA 2099 |
| PARA METHOXYBENZYL ALCOHOL |
| NSC2151 |
| 1-methoxy-4-hydroxymethyl-benzene |
| CHEBI:193647 |
| BENZENEMETHANOL,AR-METHOXY- |
| BCP26725 |
| STR00774 |
| Tox21_301115 |
| Tox21_302521 |
| BBL027471 |
| STL146342 |
| Anisyl alcohol, >=98%, FCC, FG |
| AKOS000249369 |
| anisyl alcohol (o - ,m - ,p - ) |
| Anisyl alcohol, natural, >=98%, FG |
| CS-W016493 |
| LS-2359 |
| PB47798 |
| PS-4037 |
| NCGC00248292-01 |
| NCGC00255015-01 |
| NCGC00256684-01 |
| CAS-105-13-5 |
| CAS-1331-81-3 |
| 4-METHOXY-[7-13C]-BENZYL ALCOHOL |
| 4-Methoxybenzyl alcohol, analytical standard |
| AM20020146 |
| FT-0618922 |
| FT-0671160 |
| M0107 |
| EN300-16200 |
| Anisyl alcohol 1000 microg/mL in Acetonitrile |
| D77792 |
| Q548873 |
| J-501422 |
| Z54603098 |
| F0001-0102 |
| InChI=1/C8H10O2/c1-10-8-4-2-7(6-9)3-5-8/h2-5,9H,6H2,1H |
| JandaJel(TM)-Wang, 100-200 mesh, extent of labeling: 1.0 mmol/g loading, 2 % cross-linked |
|
There are more than 10 synonyms. If you wish to see them all click here.
|