| 138-87-4 |
| cis-beta-Terpineol |
| trans-beta-Terpineol |
| p-Menth-8-en-1-ol |
| Terpinol, beta- |
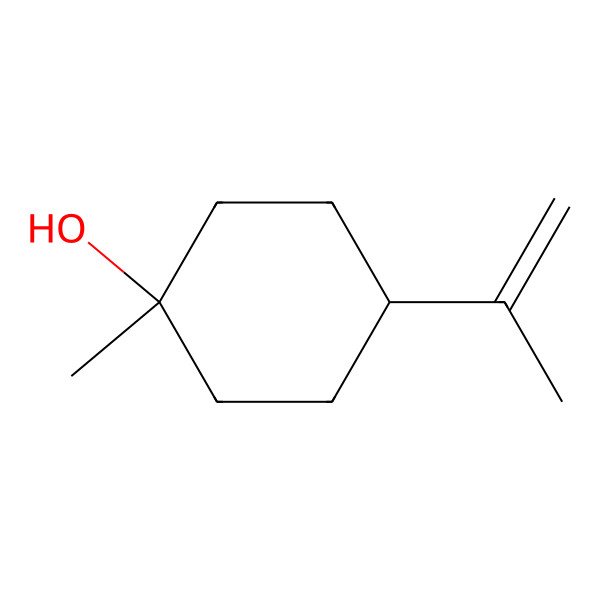
| Cyclohexanol, 1-methyl-4-(1-methylethenyl)- |
| 1-Methyl-4-(1-methylethenyl)cyclohexanol |
| cis-.beta.-Terpineol |
| trans-.beta.-terpineol |
| t-Menth-1-en-8-ol |
| 1-Methyl-4-isopropenylcyclohexan-1-ol |
| 4-Isopropenyl-1-methyl-1-cyclohexanol |
| b-terpineol |
| 1-Methyl-4-(1-methylvinyl)cyclohexan-1-ol |
| cis-|A-Terpineol |
| 1-methyl-4-(prop-1-en-2-yl)cyclohexan-1-ol |
| trans-|A-Terpineol |
| (E)-beta-Terpineol |
| FEMA No. 3564 |
| beta-Terpineol, cis- |
| beta-Terpineol, trans- |
| 4-Isopropenyl-1-methylcyclohexanol |
| Terpineol, cis-.beta.- |
| EINECS 205-342-6 |
| UNII-XS86XKC2VT |
| BRN 2205072 |
| Cyclohexanol, 1-methyl-4-(1-methylethenyl)-, cis- |
| UNII-S00R85C5ER |
| Cyclohexanol, 1-methyl-4-(1-methylethenyl)-, trans- |
| UNII-55258Z4SCW |
| AI3-00731 |
| S00R85C5ER |
| 1-methyl-4-prop-1-en-2-ylcyclohexan-1-ol |
| 7299-40-3 |
| 7299-41-4 |
| DTXSID7041209 |
| 55258Z4SCW |
| p-Menth-8-en-1-ol, stereoisomer |
| 4-06-00-00254 (Beilstein Handbook Reference) |
| beta-Terpinol |
| beta -terpineol |
| 4-isopropenyl-1-methyl-cyclohexanol |
| (E)-.beta.-terpineol |
| (Z)-.beta.-Terpineol |
| XS86XKC2VT |
| cis-p-Menth-8-en-1-ol |
| trans-p-Menth-8-en-1-ol |
| P-menth-8-en-1-ol, cis |
| SCHEMBL1245775 |
| .BETA.-TERPINEOL, CIS- |
| CHEMBL3184678 |
| DTXCID5021209 |
| SCHEMBL13895789 |
| SCHEMBL14278527 |
| SCHEMBL22882350 |
| FEMA 3564 |
| .BETA.-TERPINEOL, TRANS- |
| CHEBI:132899 |
| RUJPNZNXGCHGID-AOOOYVTPSA-N |
| RUJPNZNXGCHGID-MGCOHNPYSA-N |
| RUJPNZNXGCHGID-UHFFFAOYSA-N |
| DTXSID201317014 |
| DTXSID301316364 |
| Tox21_302378 |
| AKOS006282035 |
| HY-119963A |
| HY-119963B |
| LS-3107 |
| 4-Isopropenyl-1-methylcyclohexanol, cis |
| NCGC00255487-01 |
| CAS-138-87-4 |
| 1-methyl-4-(prop-1-en-2-yl)cyclohexanol |
| 1-Methyl-4-(1-Methylethenyl)-cyclohexanol |
| 4-Isopropenyl-1-methylcyclohexanol, trans- |
| Ciclohexanol, 1-metil-4-(1-metiletenil)- |
| CS-0089116 |
| CS-0089118 |
| FT-0692517 |
| C17517 |
| 1-Methyl-4-(1-methylethenyl)-trans-Cyclohexanol |
| TERPIN MONOHYDRATE IMPURITY B [EP IMPURITY] |
| 1-Methyl-4alpha-(1-methylethenyl)cyclohexan-1beta-ol |
| Q27261256 |
| Q27288363 |
| Q67880219 |
| 1 - methyl - 4 - (1 - methylvinyl)cyclohexan - 1 - ol |
| (1S,4S)-1-METHYL-4-(PROP-1-EN-2-YL)CYCLOHEXAN-1-OL |
|
There are more than 10 synonyms. If you wish to see them all click here.
|