| Madecassol |
| Centelase |
| 16830-15-2 |
| Asiaticosid |
| Emdecassol |
| UNII-PKO39VY215 |
| Dermatologico |
| Blastoestimulina |
| CCRIS 8995 |
| PKO39VY215 |
| EINECS 240-851-7 |
| NSC 36002 |
| NSC 166062 |
| BRN 0078195 |
| CENTELASE DERMATOLOGICO |
| CHEBI:79928 |
| 4-17-00-03627 (Beilstein Handbook Reference) |
| NSC-36002 |
| NSC-166062 |
| BA-2742 |
| Ba 2742 |
| Blastostimulina |
| Marticassol |
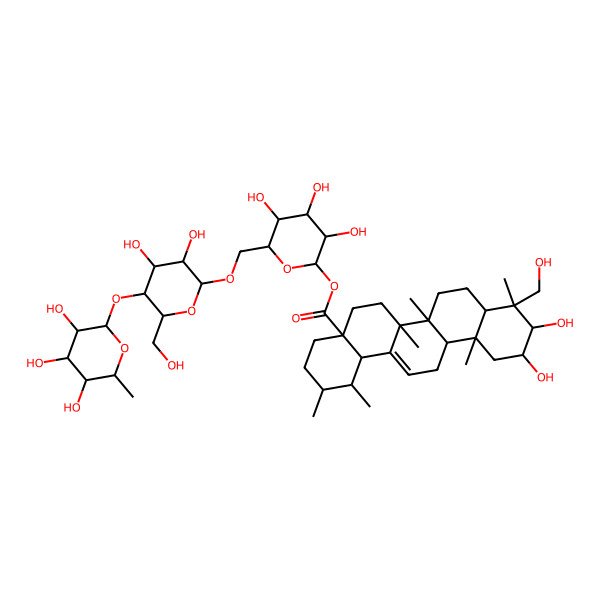
| (O-alpha-L-Rhamnopyranoxyl-(1-4)-O-beta-D-glucopyranosyl-(1-6)-O-beta-D-glucopyranosyl)-2alpha,3beta,23-trihydroxy-12-ursen-28-oat |
| 2alpha,3beta,23-Trihydroxy-urs-12-en-28-saeure(O-alpha-L-rhamnopyranosyl-(1-4)-O-beta-D-glucopyranosyl-(1-6)-O-beta-D-glucopyranosyl)ester |
| O-6-Desoxy-alpha-L-mannopyranosyl-(1-4)-O-beta-D-glucopyranosyl-(1-6)-O-beta-D-glucopyranosyl 2alpha,3beta,23-trihydroxy-12-ursen-28-at |
| ASIATICOSIDE (USP-RS) |
| ASIATICOSIDE [USP-RS] |
| O-6-Deoxy-alpha-L-mannopyranosyl-(1.4)-O-beta-D-glucopyranosyl-(1.6)-beta-D-glucopyranosyl (2alpha,3beta,4alpha)-2,3,23-trihydroxyurs-12-en-28-oate |
| Urs-12-en-28-oic acid, 2,3,23-trihydroxy-, O-6-deoxy-alpha-L-mannopyranosyl-(1-4)-O-beta-D-glucopyranosyl-(1-6)-beta-D-glucopyranosyl ester, (2alpha,3beta,4alpha)- |
| Urs-12-en-28-oic acid, 2-alpha,3-beta,23-trihydroxy-, O-6-deoxy-alpha-L-mannopyranosyl-(1-4)-o-beta-D-glucopyranosyl-(1-6)-o-beta-D-glucopyranosyl ester |
| C48-H78-O19 |
| Madecassol (TN) |
| Asiaticoside, 21 |
| ASIATICOSIDE [MI] |
| ASIATICOSIDE [INCI] |
| ASIATICOSIDE [WHO-DD] |
| CHEMBL1684590 |
| BDBM23211 |
| DTXSID30937476 |
| HY-N0439 |
| O-6-deoxy-alpha-L-mannopyranosyl-(14)-O-beta-D-glucopyranosyl-(16)-beta-D-glucopyranosyl (2alpha,3beta,4alpha)-2,3,23-trihydroxyurs-12-en-28-oate |
| Urs-12-en-28-oic acid, 2,3,23-trihydroxy-, O-6-deoxy-.alpha.-L-mannopyranosyl-(1.fwdarw.4)-O-.beta.-D-glucopyranosyl-(1.fwdarw.6)-.beta.-D-glucopyranosyl ester, (2.alpha.,3.beta.,4.alpha.)- |
| Urs-12-en-28-oic acid, 2,3,23-trihydroxy-, O-6-deoxy-.alpha.-L-mannopyranosyl-(1?4)-O-.beta.-D-glucopyranosyl-(1?6)-.beta.-D-glucopyranosyl ester, (2.alpha.,3.beta.,4.alpha.)- |
| AKOS037514554 |
| DB14081 |
| CS-0008960 |
| D07576 |
| ASIATICOSIDE (CONSTITUENT OF CENTELLA ASIATICA) |
| Q27149094 |
| ASIATICOSIDE (CONSTITUENT OF CENTELLA ASIATICA) [DSC] |
| (2.ALPHA.,3.BETA.,4.ALPHA.)-2,3,23-TRIHYDROXYURS-12-EN-28-OIC ACID O-6-DEOXY-.ALPHA.-L-MANNOPYRANOSYL-(1->4)-O-.BETA.-D-GLUCOPYRANOSYL-(1->6)-O-.BETA.-D-GLUCOPYRANOSYL ESTER |
| (2alpha,3beta)-2,3,23-Trihydroxyurs-12-en-28-oic acid, O-6-deoxy-alpha-L-mannopyranosyl-(1-4)-O-beta-D-glucopyranosyl-(1-6)-O-beta-D-glucopyranosyl Ester |
| (2alpha,3beta,4alpha)-2,3,23-TRIHYDROXYURS-12-EN-28-OIC ACID O-6-DEOXY-alpha-L-MANNOPYRANOSYL-(1->4)-O-beta-D-GLUCOPYRANOSYL-(1->6)-O-beta-D-GLUCOPYRANOSYL ESTER |
| URS-12-EN-28-OIC ACID, 2,3,23-TRIHYDROXY-, O-6-DEOXY-.ALPHA.-L-MANNOPYRANOSYL-(1->4)-O-.BETA.-D-GLUCOPYRANOSYL-(1->6)-.BETA.-D-GLUCOPYRANOSYL ESTER, (2.ALPHA.,3.BETA.,4.ALPHA.)- |
| Urs-12-en-28-oic acid, 2,3,23-trihydroxy-, O-6-deoxy-alpha-L-mannopyranosyl-(1-->4)-O-beta-D-glucopyranosyl-(1-->6)-beta-D-glucopyranosyl ester, (2alpha,3beta,4alpha)- |
| URS-12-EN-28-OIC ACID, 2,3,23-TRIHYDROXY-, O-6-DEOXY-alpha-L-MANNOPYRANOSYL-(1->4)-O-beta-D-GLUCOPYRANOSYL-(1->6)-beta-D-GLUCOPYRANOSYL ESTER, (2alpha,3beta,4alpha)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|