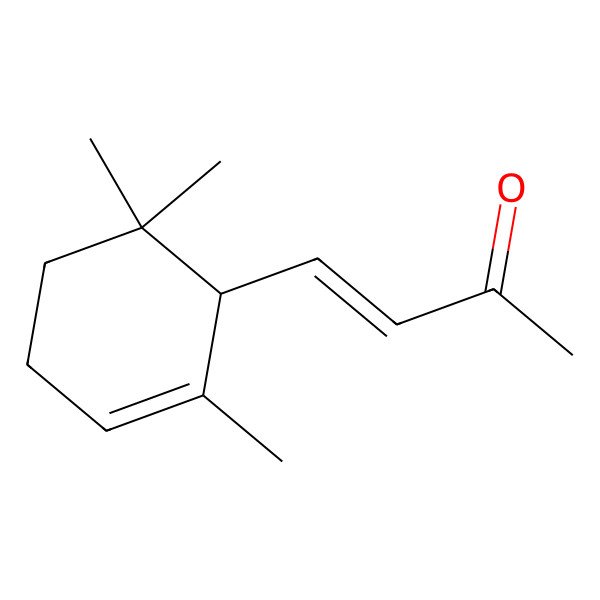
| 127-41-3 |
| (E)-alpha-Ionone |
| Iraldeine |
| alpha-Ionon |
| 4-(2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-one |
| Ionone, alpha- |
| trans-alpha-Ionone |
| alpha-(E)-ionone |
| (E)-4-(2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-one |
| Irisone |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-, (3E)- |
| a-ionone |
| alpha-Ionone (natural) |
| FEMA No. 2594 |
| alpha-Cyclocitrylideneacetone |
| Cyclocitrylideneacetone, alpha- |
| beta-lonone |
| (+-)-trans-alpha-Ionone |
| (3E)-4-(2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-one |
| .alpha.-Ionone |
| EINECS 204-841-6 |
| EINECS 250-293-6 |
| UNII-I9V075M61R |
| DTXSID0035160 |
| CHEBI:32319 |
| beta-lonone (natural) |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-, (E)- |
| I9V075M61R |
| 4-(2,6,6-Trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
| EINECS 230-010-2 |
| NSC 34560 |
| 6901-97-9 |
| (5E)-IONONE |
| .alpha.-Cyclocitrylideneacetone |
| AI3-16056 |
| DTXCID8015160 |
| HSDB 8268 |
| (E)-(1)-4-(2,6,6-Trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
| NSC34560 |
| EINECS 232-396-8 |
| EINECS 238-362-9 |
| EINECS 250-812-6 |
| 31798-12-6 |
| (-)-4-(2,6,6-Trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)- |
| (E)-4-(2,6,6-TRIMETHYL-2-CYCLOHEXEN-1-YL)-3-BUTEN-2-ONE |
| alpha-Jonone |
| -ionone |
| alpha -ionone |
| Nat. Alpha Ionone |
| MFCD00001565 |
| alpha-Ionone Bri FCC |
| IONONE, ALPHA |
| alpha-Ionone, >=90% |
| (+/-)-alpha-IONONE |
| 4,7-Megastigmadien-9-one |
| ALPHA-IONONE [FCC] |
| QP734LIN1K |
| (E)-.ALPHA.-IONONE |
| alpha-Ionone (>90per cent) |
| SCHEMBL112670 |
| .ALPHA.-IONONE [MI] |
| (E)-(+-)-4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
| CHEMBL472877 |
| .ALPHA.-IONONE [FHFI] |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-, (S-(E))- |
| alpha-Ionone, natural, >=86% |
| alpha-Ionone, analytical standard |
| (+/-)-.ALPHA.-IONONE |
| 30685-95-1 |
| Tox21_301173 |
| alpha-Ionone, technical grade, 90% |
| NSC-34560 |
| STL570068 |
| AKOS000120391 |
| AKOS016843502 |
| "4-(2,6,6-Trimethyl-cyclohex-2-e |
| (+/-)-TRANS-.ALPHA.-IONONE |
| DS-3418 |
| LS-2372 |
| alpha-Ionone, technical, >=90% (GC) |
| NCGC00164008-01 |
| NCGC00164008-02 |
| NCGC00255071-01 |
| CAS-127-41-3 |
| LS-84164 |
| CS-0149165 |
| I0076 |
| EN300-20383 |
| A889200 |
| W-108383 |
| Q27114869 |
| (e)-4-(2,6,6-trimethylcyclohex-2-enyl)but-3-en-2-one |
| 4-(2,6,6-trimethylcyclohex-2-ene-1-yl)-but-3-ene-2-one |
| (?)-4-(2,6,6-Trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
| (3E)-4-(2,6,6-Trimethyl-2-cyclohexen-1-yl)-3-buten-2-on |
| 3-buten-2-ona, 4-(2,6,6-trimetil-2-ciclohexen-1-il)-, (3e)- |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-, (-)- |
| 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-, (E)-(.+/-.)- |
| 4 - (2,6,6 - trimethylcyclohex - 2 - ene - 1 - yl) - but - 3 - ene - 2 - one |
|
There are more than 10 synonyms. If you wish to see them all click here.
|