| 98-85-1 |
| ALPHA-METHYLBENZYL ALCOHOL |
| 1-Phenylethan-1-ol |
| Styralyl alcohol |
| Styrallyl alcohol |
| Methylphenyl carbinol |
| 1-Phenethyl alcohol |
| 1-Phenylethyl alcohol |
| Methylphenylcarbinol |
| 13323-81-4 |
| Phenylmethylcarbinol |
| (1-Hydroxyethyl)benzene |
| DL-1-Phenylethanol |
| sec-Phenethyl alcohol |
| Methyl phenyl carbinol |
| (+/-)-1-Phenylethanol |
| alpha-Phenylethanol |
| 1-Phenyl-1-hydroxyethane |
| alpha-Methylbenzenemethanol |
| Ethanol, 1-phenyl- |
| DL-1-Phenethylalcohol |
| alpha-Phenethyl alcohol |
| Methanol, methylphenyl- |
| alpha-Phenylethyl alcohol |
| Styrene alcohol |
| NCI-C55685 |
| 1-Fenylethanol |
| alpha-Methyl-benzmethanol |
| Fenyl-methylkarbinol |
| Benzenemethanol, .alpha.-methyl- |
| FEMA No. 2685 |
| FEMA Number 2685 |
| Phenyl Ethanol |
| 1-Fenylethanol [Czech] |
| 1-PHENYL-ETHANOL |
| 1-phenyl ethanol |
| .alpha.-Methylbenzyl alcohol |
| NSC 25502 |
| .alpha.-Methylbenzenemethanol |
| Fenyl-methylkarbinol [Czech] |
| Benzyl alcohol, alpha-methyl- |
| Benzenemethanol, alpha-methyl- |
| CCRIS 2367 |
| (1)-alpha-Methylbenzyl alcohol |
| a-methylbenzyl alcohol |
| methyl phenyl methanol |
| .alpha.-Phenylethanol |
| Benzyl alcohol, .alpha.-methyl- |
| HSDB 4164 |
| 1-Phenyl ethyl alcohol |
| DL-sec-Phenethyl alcohol |
| EINECS 202-707-1 |
| EINECS 236-361-8 |
| UN2937 |
| .alpha.-Phenethyl alcohol |
| alpha-methyl-benzenemethanol |
| alpha-Methyl-benzyl alcohol |
| .alpha.-Phenylethyl alcohol |
| CHEBI:669 |
| UNII-E6O895DQ52 |
| AI3-02936 |
| DTXSID1020859 |
| (+/-)-alpha-Methylbenzyl alcohol |
| E6O895DQ52 |
| NSC-25502 |
| 1-PHENYLETHAN-2,2,2-D3-OL |
| Benzene-d5-methan-d-ol, a-(methyl-d3)- |
| EC 202-707-1 |
| 1-Phenylethan-1,2,2,2-d4-ol |
| alpha-Hydroxyethylbenzene |
| Benzenemethanol, ?-methyl- |
| -METHYLBENZYL ALCOHOL |
| (+/-)-1-Phenylethyl Alcohol |
| methylbenzylalcohol |
| methylbenzyl alcohol |
| 1 - phenylethanol |
| a-Hydroxyethylbenzene |
| Methylphenyl-Methanol |
| MFCD00004508 |
| 1-hydroxyethylbenzene |
| 1-phenyl-1-ethanol |
| 1-phenylethane-1-ol |
| rac.-1-phenylethanol |
| Alcohol methyl benzylic |
| m ethyl phenyl methanol |
| racemic 1-phenylethanol |
| MBA (CHRIS Code) |
| (+) alpha-phenylethanol |
| 1-Phenylethanol, 98% |
| 1-(Phenylethyl) alcohol |
| 1-Phenyl-1-ethylalcohol |
| Bencenmetanol, alfa-metil- |
| .alpha.-Hydroxyethylbenzene |
| Alpha-methyl benzyl alcohol |
| SCHEMBL2164 |
| (s)-(+)-1-phenylethanol |
| a-Methylbenzenemethanol, 9CI |
| a-Methylbenzyl alcohol, 8CI |
| racemic 1-phenylethyl alcohol |
| DTXCID50859 |
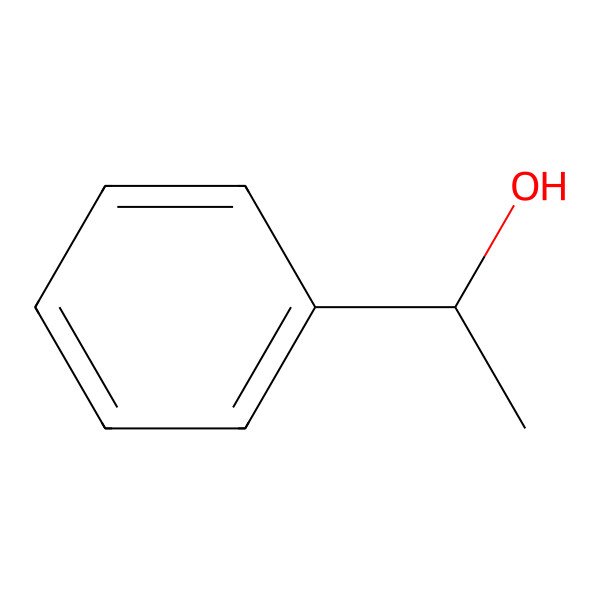
| CC(O)C1=CC=CC=C1 |
| (+/-)-sec-Phenethyl Alcohol |
| (+/-)-a-Methylbenzyl alcohol |
| CHEMBL508917 |
| WLN: QY1 & R |
| (?)-alpha-Methylbenzenemethanol |
| dl-.alpha.-Methylbenzyl alcohol |
| (+-)-alpha-methylbenzyl alcohol |
| FEMA 2685 |
| AMY3819 |
| LS-24 |
| BCP24404 |
| NSC25502 |
| Tox21_200505 |
| NA2937 |
| UN3438 |
| 1-PHENYLETHANOL, (+/-)- |
| AKOS000121326 |
| AKOS016038258 |
| SB40856 |
| UN 2937 |
| ALPHA-METHYLBENZYL ALCOHOL [FCC] |
| CAS-98-85-1 |
| ALPHA-METHYLBENZYL ALCOHOL [HSDB] |
| NCGC00248663-01 |
| NCGC00258059-01 |
| SY007833 |
| .ALPHA.-METHYLBENZYL ALCOHOL [FHFI] |
| (+/-)-.ALPHA.-METHYLBENZYL ALCOHOL |
| Benzenemethanol, .alpha.-methyl-, (.+.)- |
| Benzyl alcohol, .alpha.-methyl-, (.+.)- |
| FT-0605197 |
| FT-0628703 |
| FT-0633624 |
| FT-0647462 |
| M0163 |
| Benzenemethanol, .alpha.-methyl-, (.+-.)- |
| EN300-21267 |
| alpha-Methylbenzyl alcohol [UN2937] [Poison] |
| alpha-Methylbenzyl alcohol, >=99%, FCC, FG |
| C07112 |
| alpha-Methylbenzyl alcohol [UN2937] [Poison] |
| (R)-(+)-1-Phenylethanol, 99%, ChiraSelect(TM) |
| Q3740715 |
| W-100061 |
| Z104494956 |
| 5-(1-HYDROXY-2,6,6-TRIMETHYL-4-OXOCYCLOHEX-2-ENYL)-3-METHYLPENTA-2,4-DIENOICACID |
|
There are more than 10 synonyms. If you wish to see them all click here.
|