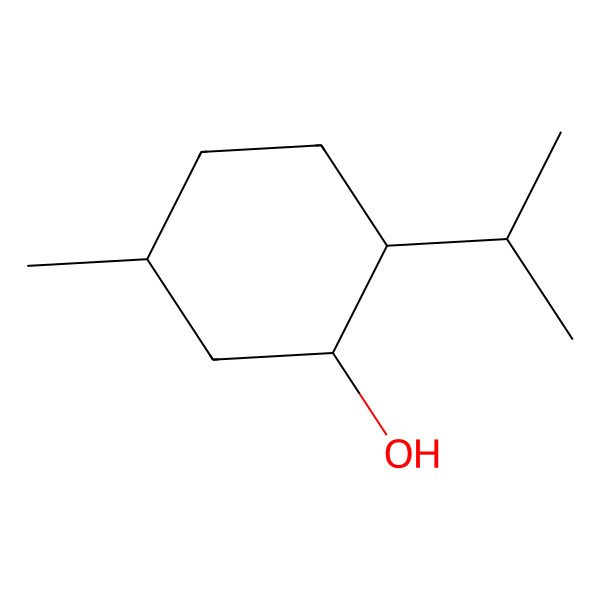
| 15356-60-2 |
| d-Menthol |
| (1S,2R,5S)-(+)-Menthol |
| (1S,2R,5S)-Menthol |
| Cyclohexanol, 5-methyl-2-(1-methylethyl)-, (1S,2R,5S)- |
| (1S,2R,5S)-2-Isopropyl-5-methylcyclohexanol |
| Hexahydrothymol |
| Menthol, (+)- |
| (1S,2R,5S)-5-methyl-2-propan-2-ylcyclohexan-1-ol |
| (1S,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexan-1-ol |
| Menthol, (1S,3S,4R)-(+)- |
| UNII-C6B1OE8P3W |
| C6B1OE8P3W |
| rac-Menthol |
| DTXSID8029733 |
| CHEBI:76306 |
| (+)-(1S,2R,5S)-menthol |
| (+)-(1S,3S,4R)-menthol |
| p-Menthan-3-ol |
| EINECS 239-387-8 |
| MFCD00062983 |
| (1S,2R,5S)-5-methyl-2-(1-methylethyl)cyclohexanol |
| 89-78-1 |
| EC 239-387-8 |
| Menthol, racemic |
| 3-p-Menthanol |
| D - menthol |
| Racementhol [BAN:INN] |
| (+)-p-Menthan-3-ol |
| Fisherman's friend lozenges |
| (1R,2S,5R)-Menthol |
| Menthol, cis-1,3,trans-1,4- |
| Fancol menthol |
| Cyclohexanol, 5-methyl-2-(1-methylethyl)-, (1.alpha.,2.beta.,5.alpha.)- |
| (DL)-Menthol |
| (+) - menthol |
| (.+/-.)-Menthol |
| (1S)-(+)-menthol |
| (+/-)-Menthol racemic |
| MENTHOL, D- |
| Ciclohexanol, 5-metil-2-(1-metiletil)-, (1r, 2s, 5r)-rel- |
| SCHEMBL521946 |
| DTXCID409733 |
| GTPL2471 |
| CHEMBL2106989 |
| (1s, 2r, 5s)-(+)-menthol |
| Menthol, (1S,3S,4R)-()- |
| Tox21_201043 |
| AKOS006281173 |
| CS-W017993 |
| DB11344 |
| HY-W017277 |
| NCGC00248905-01 |
| NCGC00258596-01 |
| AS-69562 |
| (1S,2R,5S)-(+)-Menthol, 99% |
| CAS-15356-60-2 |
| M0826 |
| M3170 |
| EN300-92162 |
| A815982 |
| (1S,2R,5S)-2-Isopropyl-5-methylcyclohexan-1-ol |
| (1S,2R,5S)-5-methyl-2-propan-2-yl-1-cyclohexanol |
| Q27084428 |
| (1S,2R,5S)-5-methyl-2-propan-2-yl-cyclohexan-1-ol |
| F8889-8741 |
| Z1198149783 |
| 5.alpha.-Methyl-2.beta.-(1.alpha.-methylethyl)cyclohexanol |
| Cyclohexanol,5-methyl-2-(1-methylethyl)-,(1S,2R,5S)- |
| 2-Isopropyl-5-methylcyclohexanol, (1.alpha.,2.beta.,5.alpha.)- |
| 5-Methyl-2-(1-methylethyl)cyclohexanol, (1.alpha.,2.beta.,5.alpha.)- |
| Cyclohexanol, 2-isopropyl-5-methyl-, (1.alpha.,2.beta.,5.alpha.)- |
| Cyclohexanol, 5-methyl-2-(1-methylethyl)-, (1S-(1alpha,2beta,5alpha))- |
| (+)-Menthol, puriss. p.a., terpene standard for GC, >=99.0% (sum of enantiomers, GC) |
| Cyclohexanol, 5-methyl-2-(1-methylethyl)-, [1S-(1.alpha.,2.beta.,5.alpha.)]- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|