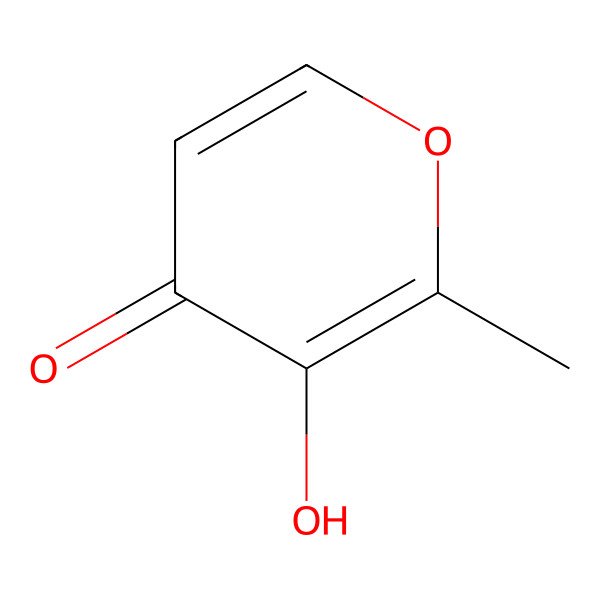
| 118-71-8 |
| 3-Hydroxy-2-methyl-4-pyrone |
| 3-Hydroxy-2-methyl-4H-pyran-4-one |
| Larixinic acid |
| Palatone |
| Larixic acid |
| Talmon |
| Vetol |
| Veltol |
| Corps praline |
| 3-hydroxy-2-methylpyran-4-one |
| 4H-Pyran-4-one, 3-hydroxy-2-methyl- |
| 2-Methyl pyromeconic acid |
| 2-Methylpyromeconic acid |
| 2-Methyl-3-hydroxy-4-pyrone |
| 2-Methyl-3-hydroxypyrone |
| Maltol (natural) |
| 3-Hydroxy-2-methyl-gamma-pyrone |
| 2-Methyl-3-oxy-gamma-pyrone |
| FEMA No. 2656 |
| 3-Hydroxy-2-methylpyrone |
| 3-Hydroxy-2-methyl-pyran-4-one |
| MFCD00006578 |
| NSC 2829 |
| Maltol [NF] |
| CCRIS 3467 |
| 2-methyl-3-hydroxy-4-pyranone |
| 3-Hydroxy-2-methyl-4-pyranone |
| CHEBI:69438 |
| NSC2829 |
| 3-Hydroxy-2-methyl-1,4-pyrone |
| Ins no.636 |
| NSC-2829 |
| 3-Hydroxy-2-methyl-.gamma.-pyrone |
| EINECS 204-271-8 |
| NSC-404458 |
| Ins-636 |
| BRN 0112169 |
| UNII-3A9RD92BS4 |
| AI3-18547 |
| MLS000069412 |
| 3A9RD92BS4 |
| DTXSID0025523 |
| 5-Hydroxy-6-methyl-4H-pyran-4-one |
| Maltol (3-Hydroxy-2-methyl-4-pyrone) |
| E636 |
| SMR000059093 |
| E-636 |
| 5-18-01-00114 (Beilstein Handbook Reference) |
| DTXCID305523 |
| WLN: T6O DVJ B1 CQ |
| CAS-118-71-8 |
| Methylmaltol |
| methyl maltol |
| Laricinic acid |
| PIROMATOL |
| NATURAL MALTOL |
| MALTOL, NATURAL |
| Spectrum_001419 |
| Opera_ID_338 |
| SpecPlus_000443 |
| MALTOL [FHFI] |
| MALTOL [INCI] |
| MALTOL [FCC] |
| MALTOL [USP-RS] |
| Spectrum2_001795 |
| Spectrum3_001351 |
| Spectrum4_001871 |
| Spectrum5_000462 |
| MALTOL [II] |
| MALTOL [MI] |
| MALTOL [MART.] |
| bmse000538 |
| Maltol, analytical standard |
| SCHEMBL4815 |
| 3-Hydroxy-2-pyran-4-one |
| BSPBio_003161 |
| KBioGR_002365 |
| KBioSS_001899 |
| SPECTRUM310025 |
| MLS001424145 |
| MLS002415738 |
| 3-Hydroxy-2-methyl-g-pyrone |
| CHEMBL31422 |
| DivK1c_006539 |
| 3-hydroxy-2-methylpyr-4-one |
| SPBio_001749 |
| QSPL 180 |
| 2-Methyl-3-oxy-.gamma.-pyrone |
| 3-hydroxy-2-methyl-4-oxopyrane |
| 3-hydroxyl-2-methyl-4-pyranone |
| FEMA 2656 |
| HSDB 8320 |
| KBio1_001483 |
| KBio2_001899 |
| KBio2_004467 |
| KBio2_007035 |
| KBio3_002381 |
| XPCTZQVDEJYUGT-UHFFFAOYSA- |
| 3-hydroxy-2-methyl-gamma -pyrone |
| HMS2052K09 |
| HMS3394K09 |
| KUC106764N |
| STR01642 |
| Tox21_202215 |
| Tox21_300118 |
| BBL011669 |
| BDBM50227434 |
| CCG-38443 |
| LS-602 |
| Maltol, natural, >=98.5%, FG |
| NSC404458 |
| s4940 |
| STK801686 |
| 2-methyl-3-hydroxy-4H-pyran-4-one |
| Maltol, >=99.0%, FCC, FG |
| 3-Hydroxy-2-methyl-4-pyrone, 99% |
| AKOS005607790 |
| 3-Hydroxy-2-Methyl-4-pyrone, natural |
| CS-W013504 |
| HY-W012788 |
| NC00350 |
| PS-4578 |
| SDCCGMLS-0066563.P001 |
| 4H-piran-4-ona, 3-hidroxi-2-metil- |
| 4-(a-d-Glucopyranosido)-a-glucopyranose |
| NCGC00091223-01 |
| NCGC00091223-02 |
| NCGC00091223-03 |
| NCGC00091223-04 |
| NCGC00091223-05 |
| NCGC00178231-01 |
| NCGC00254046-01 |
| NCGC00259764-01 |
| BP-11468 |
| KSC-11-228-8 |
| NCI60_002320 |
| SY011358 |
| PYR-4-ONE, 3-HYDROXY-2-METHYL- |
| 3 - hydroxy - 2 - methyl - 4 - pyrone |
| AM20080119 |
| FT-0615804 |
| M0673 |
| EN300-93557 |
| A804081 |
| Q420648 |
| SR-01000712383 |
| SR-01000712383-3 |
| W-108539 |
| BRD-K40619305-001-12-1 |
| Z1255382969 |
| Maltol, United States Pharmacopeia (USP) Reference Standard |
| InChI=1/C6H6O3/c1-4-6(8)5(7)2-3-9-4/h2-3,8H,1H3 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|