| 489-86-1 |
| Champacol |
| Guaiac alcohol |
| Champaca camphor |
| (-)-Guaiol |
| Guai-1(5)-en-11-ol |
| GUAIOL(-) |
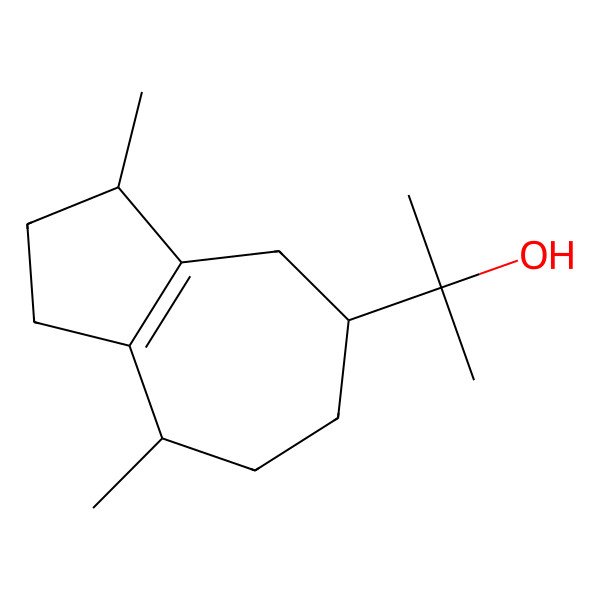
| 2-[(3S,5R,8S)-3,8-dimethyl-1,2,3,4,5,6,7,8-octahydroazulen-5-yl]propan-2-ol |
| UNII-I7WP008A91 |
| CHEBI:5552 |
| I7WP008A91 |
| EINECS 207-702-8 |
| NSC 19941 |
| NSC-19941 |
| NSC19941 |
| 5-Azulenemethanol, 1,2,3,4,5,6,7,8-octahydro-.alpha.,.alpha.,3,8-tetramethyl-, (3S,5R,8S)- |
| 5-Azulenemethanol, 1,2,3,4,5,6,7,8-octahydro-alpha,alpha,3,8-tetramethyl-, (3S,5R,8S)- |
| [3S-(3alpha,5alpha,8alpha)]-1,2,3,4,5,6,7,8-octahydro-alpha,alpha,3,8-tetramethyl-5-azulenemethanol |
| 2-((3S,5R,8S)-3,8-dimethyl-1,2,3,4,5,6,7,8-octahydroazulen-5-yl)propan-2-ol |
| 2-((3S,8S)-1,2,3,4,5,6,7,8-Octahydro-3,8-dimethylazulen-5-yl)propan-2-ol |
| Champacol; Guaiac alcohol |
| (3S-(3alpha,5alpha,8alpha))-1,2,3,4,5,6,7,8-octahydro-alpha,alpha,3,8-tetramethyl-5-azulenemethanol |
| (?)-Guaiol |
| Spectrum3_001870 |
| GUAIOL [MI] |
| (-)-Guaiol, 97% |
| BSPBio_003320 |
| SCHEMBL114056 |
| SPECTRUM1800009 |
| 3,8-dimethyl-5-(alpha-hydroxyisopropyl)-delta-9-octahydroazulene |
| FEMA No. 2534 |
| CHEMBL226915 |
| HSDB 3437 |
| KBio3_002822 |
| (-)-Guaiol, analytical standard |
| DTXSID40883399 |
| (3S,5R,8S)-1,2,3,4,5,6,7,8-Octahydro-alpha,-alpha,3,8-tetramethyl-5-azulenemethanol |
| 2 - ((3S,8S) - 1,2,3,4,5,6,7,8 - octahydro - 3,8 - dimethylazulen - 5 - yl)propan - 2 - ol |
| 5-Azulenemethanol, 1,2,3,4,5,6,7,8-octahydro- ?,?,3,8-tetramethyl-, [3S-(3?,5?,8?)]- |
| 5-Azulenemethanol, 1,2,3,4,5,6,7,8-octahydro-alpha,alpha,3,8-tetramethyl-, (3S-(3alpha,5alpha,8alpha))- |
| HY-N3980 |
| MFCD00043336 |
| AKOS040760436 |
| Guaiol 100 microg/mL in Acetonitrile |
| CCG-208248 |
| LMPR0103410007 |
| NCGC00178142-01 |
| LS-73185 |
| MS-23248 |
| CS-0024560 |
| C09676 |
| E88593 |
| SR-05000002468 |
| Q5613321 |
| SR-05000002468-1 |
| (3R,6S,10S)-6,10,a,a-Tetramethylbicyclo[5.3.0]dec-1(7)-ene-3-methanol |
| 2-(3,8-dimethyl-1,2,3,4,5,6,7,8-octahydro-azulen-5-yl)-propan-2-ol |
| (3R,6S,10S)-6,10,alpha,alpha-Tetramethylbicyclo[5.3.0]dec-1(7)-ene-3-methanol |
| (3S,5R,8S)-1,2,3,4,5,6,7,8-OCTAHYDRO-alpha,alpha3,8-TETRAMETHYL-5-AZULENEMETHANOL |
| (3S,5R,8S)-1,2,3,4,5,6,7,8-OCTAHYDRO-.ALPHA.,.ALPHA.3,8-TETRAMETHYL-5-AZULENEMETHANOL |
| 5-Azulenemethanol,2,3,4,5,6,7,8-octahydro-.alpha.,.alpha.,3,8-tetramethyl-, [3S-(3.alpha.,5.alpha.,8.alpha.)]- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|