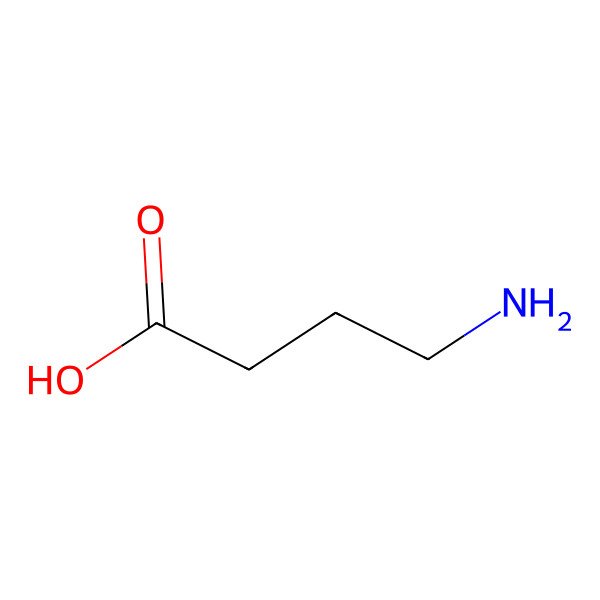
| 4-Aminobutanoic acid |
| gamma-aminobutyric acid |
| GABA |
| 56-12-2 |
| Piperidic acid |
| Piperidinic acid |
| Aminalon |
| Gaballon |
| Gammalon |
| Gammalone |
| Gammasol |
| Mielogen |
| Mielomade |
| Gamarex |
| Gammar |
| butanoic acid, 4-amino- |
| Reanal |
| Butyric acid, 4-amino- |
| gamma-amino-n-butyric acid |
| Aminobutyric acid |
| Immu-G |
| gamma-Aminobutanoic acid |
| Gamastan |
| Gammagee |
| Gamulin |
| .gamma.-Aminobutyric acid |
| omega-Aminobutyric acid |
| Aminalone |
| gamma-Amino butyric acid |
| gamma-Aminobuttersaeure |
| gamma-aminobutyrate |
| 3-Carboxypropylamine |
| GAMMA-AMINO-BUTANOIC ACID |
| 4-aminobutyrate |
| 4-aminobutanoate |
| 4-amino-butanoic acid |
| gamma Aminobutyric acid |
| DF 468 |
| CCRIS 3721 |
| 4-Aminobutyricacid |
| 4Abu |
| 4-amino butyric acid |
| 4-Amino-butyric acid |
| NSC 27418 |
| Gamma-aminobutyric acid [JAN] |
| EPA Pesticide Chemical Code 030802 |
| GAMMA(AMINO)-BUTYRIC ACID |
| UNII-2ACZ6IPC6I |
| 2ACZ6IPC6I |
| Aminobutyric acid, gamma- |
| Factor I |
| AI3-26812 |
| Aminobutanoic acid |
| .gamma.-Amino-butyric acid |
| EINECS 200-258-6 |
| NSC-27418 |
| g-Aminobutyric acid |
| .gamma.-Amino-N-butyric acid |
| DTXSID6035106 |
| CHEBI:16865 |
| gamma-Aminobuttersaeure [German] |
| gamma-Aminobutryic acid |
| 4-amino-n-butyric acid |
| 4-NH2-but |
| 4-NH3-but |
| Acide amino-4- butyrique [French] |
| CHEMBL96 |
| Acide amino-4- butyrique |
| .gamma.-Aminobutanoic acid |
| [3H]GABA |
| MLS000028505 |
| gamma-Aminobutyric acid (JAN) |
| 20-79-1 |
| DTXCID4015106 |
| 4-Aminobutyric-2,2-D2 acid |
| Butanoic acid, 4-amino- (9CI) |
| NSC27418 |
| NSC32044 |
| NSC45460 |
| NSC51295 |
| 4-Aminobutylate |
| Butyric acid, 4-amino- (7CI,8CI) |
| NCGC00015043-07 |
| SMR000058285 |
| 4-AMINO-BUTYRATE |
| WLN: Z3VQ |
| BUTANOIC ACID,4-AMINO |
| GAMMA-AMINOBUTYRIC ACID (MART.) |
| GAMMA-AMINOBUTYRIC ACID [MART.] |
| GAMMA-AMINOBUTYRIC ACID (USP-RS) |
| GAMMA-AMINOBUTYRIC ACID [USP-RS] |
| Chemical Name: .gamma.-Aminobutanoic acid |
| 4 Aminobutyric Acid |
| VIGABATRIN IMPURITY D (EP IMPURITY) |
| VIGABATRIN IMPURITY D [EP IMPURITY] |
| 4 Aminobutanoic Acid |
| SR-01000075618 |
| aminobutyrate |
| Immuglobin |
| Piperidate |
| Piperidinate |
| SODIUM ALENDRONATE TRIHYDRATE IMPURITY A (EP IMPURITY) |
| SODIUM ALENDRONATE TRIHYDRATE IMPURITY A [EP IMPURITY] |
| w-Aminobutyrate |
| 4-Aminobutyric |
| Gamma aminobutyrate |
| omega-Aminobutyrate |
| Gammalon (TN) |
| w-Aminobutyric acid |
| y-Aminobutyric acid |
| ?-Aminobutyric acid |
| MFCD00008226 |
| gamma-Aminobuttersaure |
| Vigabatrin impurity D |
| 4- aminobutyric acid |
| gamma-aminobutyric-acid |
| gam.-Aminobutyric acid |
| Spectrum_000049 |
| Tocris-0344 |
| Aminobutyric acid,-4- |
| gamma-amino-butyric acid |
| Acide amino-4-butyrique |
| Opera_ID_1152 |
| Spectrum2_001208 |
| Spectrum3_001385 |
| Spectrum4_000809 |
| Spectrum5_001425 |
| Lopac-A-2129 |
| Vigabatrin EP Impurity D |
| .omega.-Aminobutyric acid |
| cid_119 |
| Biomol-NT_000230 |
| bmse000340 |
| bmse000820 |
| bmse000871 |
| D0C9XA |
| D0D3ID |
| SCHEMBL4878 |
| Lopac0_000005 |
| Oprea1_584567 |
| BSPBio_002970 |
| Gamma-Aminobutyric Acid,(S) |
| KBioGR_001297 |
| KBioSS_000429 |
| DivK1c_000616 |
| NH2-(CH2)3-COOH |
| SPECTRUM1500678 |
| SPBio_000996 |
| GTPL1067 |
| GTPL5410 |
| SGCUT00121 |
| AMINOBUTYRIC ACID [INCI] |
| BDBM24183 |
| HMS501O18 |
| KBio1_000616 |
| KBio2_000429 |
| KBio2_002997 |
| KBio2_005565 |
| KBio3_002190 |
| gamma-Aminobutyric acid, >=99% |
| NINDS_000616 |
| HMS1921C06 |
| HMS2232P09 |
| HMS3260A11 |
| Pharmakon1600-01500678 |
| 4-AMINOBUTYRIC ACID [FHFI] |
| BCP34675 |
| HY-N0067 |
| STR01523 |
| to_000021 |
| 4-amino-n-[2,3-3H]butyric acid |
| Tox21_110071 |
| Tox21_500005 |
| BBL008590 |
| CCG-38515 |
| GSK683699 |
| HB0882 |
| LMFA01100039 |
| NSC-32044 |
| NSC-45460 |
| NSC-51295 |
| NSC757426 |
| PDSP1_001275 |
| PDSP2_001259 |
| s4700 |
| STK301748 |
| AKOS000119660 |
| CS-W020704 |
| DB02530 |
| KS-5273 |
| LP00005 |
| NSC-757426 |
| SDCCGSBI-0049994.P004 |
| .GAMMA.-AMINOBUTYRIC ACID [MI] |
| CAS-56-12-2 |
| IDI1_000616 |
| GAMMA-AMINOBUTYRIC ACID [WHO-DD] |
| NCGC00015043-01 |
| NCGC00015043-02 |
| NCGC00015043-03 |
| NCGC00015043-04 |
| NCGC00015043-05 |
| NCGC00015043-06 |
| NCGC00015043-08 |
| NCGC00015043-09 |
| NCGC00015043-10 |
| NCGC00015043-14 |
| NCGC00024546-01 |
| NCGC00024546-02 |
| NCGC00024546-03 |
| NCGC00024546-04 |
| NCGC00024546-05 |
| NCGC00024546-06 |
| NCGC00260690-01 |
| BP-21452 |
| LS-47805 |
| gamma-Aminobutyric acid, BioXtra, >=99% |
| SBI-0049994.P003 |
| gamma-Aminobutyric acid, analytical standard |
| A0282 |
| AM20100372 |
| EU-0100005 |
| FT-0617569 |
| EN300-19823 |
| gamma-Aminobutyric acid (4-Aminobutyric acid) |
| 4-aminobutanoic acid (gamma-aminobutyric acid) |
| A 2129 |
| A-5290 |
| A14934 |
| C00334 |
| D00058 |
| D70585 |
| Vigabatrin impurity, .gamma.-aminobutyric acid- |
| AB00052155_12 |
| L000262 |
| Q210021 |
| SR-01000075618-1 |
| SR-01000075618-3 |
| SR-01000075618-6 |
| SR-01000075618-7 |
| BRD-K77245796-001-19-2 |
| BUTANOIC ACID,4-AMINO 4-AMINO,BUTYRIC ACID |
| F2191-0196 |
| Z104475620 |
| C5C3DC27-C105-4D18-8A52-F51978B32D24 |
| gamma-Aminobutyric acid pound>>(c)(3/4)?Aminobutyric acid (GABA) |
| gamma-Aminobutyric acid, certified reference material, TraceCERT(R) |
| InChI=1/C4H9NO2/c5-3-1-2-4(6)7/h1-3,5H2,(H,6,7 |
| VIGABATRIN IMPURITY, gamma-AMINOBUTYRIC ACID-(USP IMPURITY) |
| Vigabatrin impurity D, European Pharmacopoeia (EP) Reference Standard |
| VIGABATRIN IMPURITY, .GAMMA.-AMINOBUTYRIC ACID- [USP IMPURITY] |
| gamma-Aminobutyric acid, United States Pharmacopeia (USP) Reference Standard |
|
There are more than 10 synonyms. If you wish to see them all click here.
|