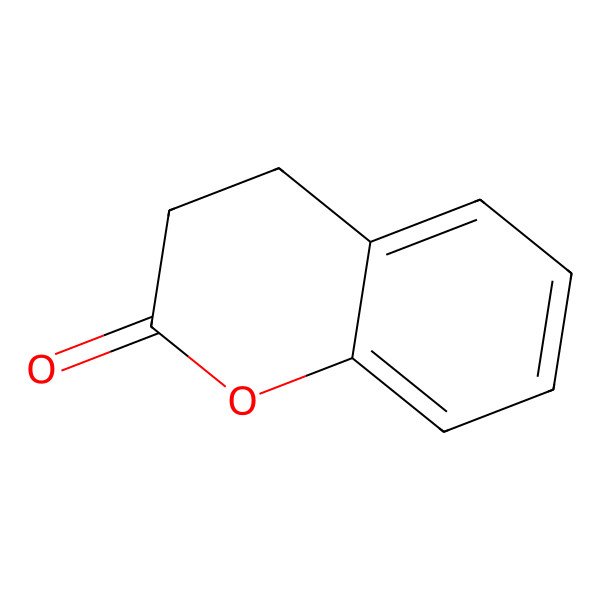
| 3,4-dihydrocoumarin |
| 119-84-6 |
| hydrocoumarin |
| chroman-2-one |
| Benzodihydropyrone |
| melilotin |
| 2-chromanone |
| melilotol |
| 1,2-benzodihydropyrone |
| melilotic lactone |
| Melilotine |
| Oxochroman |
| 3,4-Dihydro-2H-1-benzopyran-2-one |
| Melilotic acid lactone |
| Chroman, 2-oxo- |
| 2H-1-Benzopyran-2-one, 3,4-dihydro- |
| Benzopyranone, dihydro- |
| Dihydrobenzopyrone |
| Coumarin, 3,4-dihydro- |
| Usaf do-12 |
| 2-Hydroxydihydrocinnamic acid lactone |
| Meliotine |
| NCI-C55890 |
| 3,4-dihydrochromen-2-one |
| 3,4-Dyhydrocoumarin |
| 3,4-Dihydro-1-benzopyran-2-one |
| 3,4-dihydro-2H-chromen-2-one |
| o-hydroxydihydrocinnamic acid lactone |
| FEMA No. 2381 |
| 2-oxo-chroman |
| Hydrocinnamic acid, o-hydroxy-, delta-lactone |
| Hydroxydihydrocinnamic acid lactone, o- |
| CCRIS 5803 |
| o-hydroxyhydrocinnamic acid delta-lactone |
| HSDB 4333 |
| o-hydroxyhydrocinnamic acid lactone |
| EINECS 204-354-9 |
| NSC 10121 |
| UNII-NM5K1Y1BT2 |
| BRN 0004584 |
| NM5K1Y1BT2 |
| CHEMBL89306 |
| COUMARIN,3,4-DIHYDRO |
| AI3-03425 |
| DTXSID2020474 |
| CHEBI:16151 |
| NSC10121 |
| NSC-10121 |
| hydrocinnamic acid, o-hydroxy-,lactone |
| Hydrocinnamic acid, o-hydroxy-, .delta.-lactone |
| 5-17-10-00013 (Beilstein Handbook Reference) |
| DTXCID00474 |
| CAS-119-84-6 |
| dihydrocumarin |
| dihydrocoumarine |
| hydrocoumar in |
| 2-Oxochroman |
| Melilotin?? |
| Dihydrobenzenopyrone |
| Hydrocoumarin, 8CI |
| Dihydro-Benzopyranone |
| MFCD00006881 |
| 2,3-dihydrocoumarin |
| DIHIDROCUMARINA |
| 3,4-Dihydrocumarine |
| Coumarin,4-dihydro- |
| Melilotin (coumarin) |
| 3,4-dihydrocoumarine |
| 3,4 -dihydrocoumarin |
| 3,4-Dihydroxycoumarin |
| 3,4-Dihydro-Coumarin |
| Dihydrocoumarin, 99% |
| 3, 4- dihydrocoumarin |
| MELILOTIN [HSDB] |
| bmse000412 |
| SCHEMBL28795 |
| MLS002454372 |
| 2-Hydroxyhydrocinnamic lactone |
| DIHYDROCOUMARIN [FHFI] |
| DIHYDROCOUMARIN [INCI] |
| LS-9 |
| WLN: T66 BOVT & J |
| FEMA 2381 |
| HMS2268K22 |
| Dihydrocoumarin, analytical standard |
| HY-N1926 |
| Hydrocinnamic acid, .delta.-lactone |
| Tox21_202137 |
| Tox21_302745 |
| 2H-1-Benzopyran-2-one,4-dihydro- |
| BBL027621 |
| BDBM50146070 |
| s9379 |
| STK801851 |
| Dihydrocoumarin, >=99%, FCC, FG |
| 1,2-BENZODIHYDROPYRONE [FCC] |
| AKOS000277415 |
| Benzo-DiHydro Pyrone/Di Hydro Coumarin |
| CCG-266187 |
| 2h-1-benzopiran-2-ona, 3,4-dihidro- |
| NCGC00091491-01 |
| NCGC00091491-02 |
| NCGC00256356-01 |
| NCGC00259686-01 |
| AC-34197 |
| NCI60_000035 |
| SMR000112325 |
| VS-08570 |
| 2H-1-Benzopyran-2-one, 3, 4-dihydro- |
| CS-0018237 |
| D1223 |
| FT-0614320 |
| Dihydrocoumarin 100 microg/mL in Acetonitrile |
| EN300-49183 |
| C02274 |
| D70234 |
| A892403 |
| O-HYDROXY-HYDROCINNAMIC ACID-.DELTA.-LACTONE |
| W-108495 |
| Q27098403 |
| Z586249466 |
| InChI=1/C9H8O2/c10-9-6-5-7-3-1-2-4-8(7)11-9/h1-4H,5-6H |
|
There are more than 10 synonyms. If you wish to see them all click here.
|