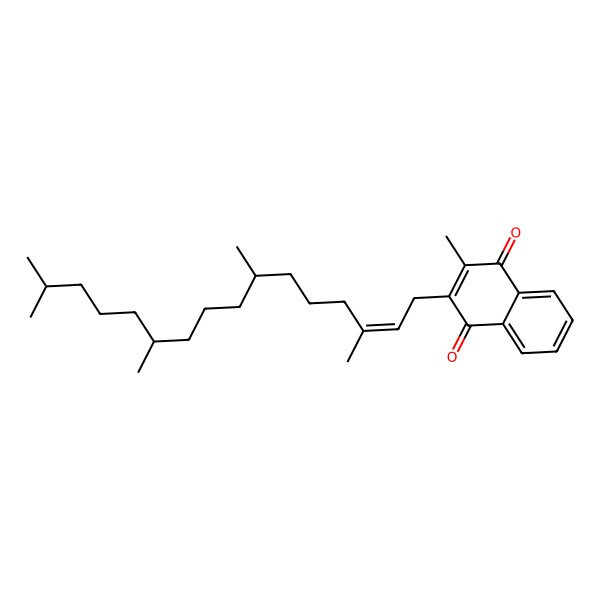
| VITAMIN K |
| 81818-54-4 |
| Kephton |
| Kinadion |
| 2-methyl-3-[(E)-3,7,11,15-tetramethylhexadec-2-enyl]naphthalene-1,4-dione |
| Aqua mephyton |
| Vitamin K1(20) |
| .alpha.-Phylloquinone |
| Mono-Kay |
| Combinal K1 |
| Kativ N |
| K-Ject |
| Vitamin K semiquinone radical |
| Phythyl-menadion |
| 2',3'-trans-Vitamin K1 |
| EINECS 234-408-7 |
| EINECS 279-833-9 |
| 2-methyl-3-(3,7,11,15-tetramethylhexadec-2-en-1-yl)naphthalene-1,4-dione |
| VitaminK1-18O(Mixtureof1-18Oand4-18O) |
| 2-methyl-3-(3,7,11,15-tetramethylhexadec-2-enyl)naphthalene-1,4-dione |
| 10485-69-5 |
| Aqua-Mephytin |
| Phyllochinon [German] |
| (E)-2-Methyl-3-(3,7,11,15-tetramethylhexadec-2-en-1-yl)naphthalene-1,4-dione |
| (E)-2-Methyl-3-(3,7,11,15-tetramethylhexadec-2-en-1-yl)naphthalene-1,4-dione1 |
| 2-methyl-3-[(2E)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-1,4-dihydronaphthalene-1,4-dione |
| Fitomenadione [DCIT] |
| Vitamin K1 (VAN) |
| 2581046-19-5 |
| 27696-10-2 |
| SMR000059144 |
| Phytonadione [USAN:JAN] |
| Fitomenadiona [INN-Spanish] |
| Phytomenadionum [INN-Latin] |
| C31H46O2 |
| NSC270681 |
| Vitamina K |
| 2-Methyl-3-(3,7,11,15-tetramethylhexadec-2-enyl)-1,4-naphthoquinone |
| 2-Methyl-3-phythyl-1,4-naphthochinon |
| C31-H46-O2 |
| 2-methyl-3-[(2E)-3,7,11,15-tetramethylhexadec-2-en-1-yl]naphthalene-1,4-dione |
| NCGC00159423-02 |
| Vitamin K substances |
| Vitamin K (generic) |
| 2-Methyl-3-phytyl-1,4-naphthochinon [German] |
| starbld0012504 |
| D07QPA |
| Vitamin K1 (Phytonadione) |
| Phytonadione (JP17/USP) |
| MLS001332659 |
| MLS001332660 |
| CHEMBL520156 |
| 2-Methyl-3-(3,7,11,15-tetramethyl-2-hexadecenyl)-1,4-naphthalened- ione |
| 2-Methyl-3-[(2E)-3,7,11,15-tetramethyl-2-hexadecenyl]naphthoquinone # |
| CHEBI:94399 |
| [r-[r*,R*-(E)]]-2-Methyl-3-(3,7,11,15-tetramethyl-2-hexadecenyl)-1,4-napthalenedione |
| [r-[r*,R*-(E)]]-2-Methyl-3-(3-7,11,15-tetramethyl-2-hexadecenyl)-1,4-naphthalenedione |
| HMS2232C17 |
| BCP23822 |
| BBL036678 |
| STL559057 |
| AKOS024284357 |
| 3-((2E)-3,7,11,15-tetramethylhexadec-2-enyl)-2-methylnaphthalene-1,4-dione |
| NCGC00186656-01 |
| LS-94617 |
| VS-13623 |
| P0642 |
| C02059 |
| D00148 |
| 1, 2-methyl-3-(3,7,11,15-tetramethyl-2-hexadecenyl)- |
| 2-(3,7,11,15-Tetramethyl-2-hexadecenyl)-3-methyl-1,4-naphthoquinone |
| 2-Methyl-3-(3,11,15-tetramethyl-2-hexadecenyl)-1,4-naphthalenedione |
| 1, 2-methyl-3-(3,7,11,15-tetramethyl-2-hexadecenyl)-, [R-[R*,R*-(E)]]- |
| 2-Methyl-3-[(2Z)-3,7,11,15-tetramethyl-2-hexadecenyl]-1,4-naphthoquinone |
| 1,4-Naphthalenedione, 2-methyl-3-(3,7,11,15-tetramethyl-2-hexadecenyl)-, radical ion(1-), (R-(R*,R*-(E)))- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|