| 121-79-9 |
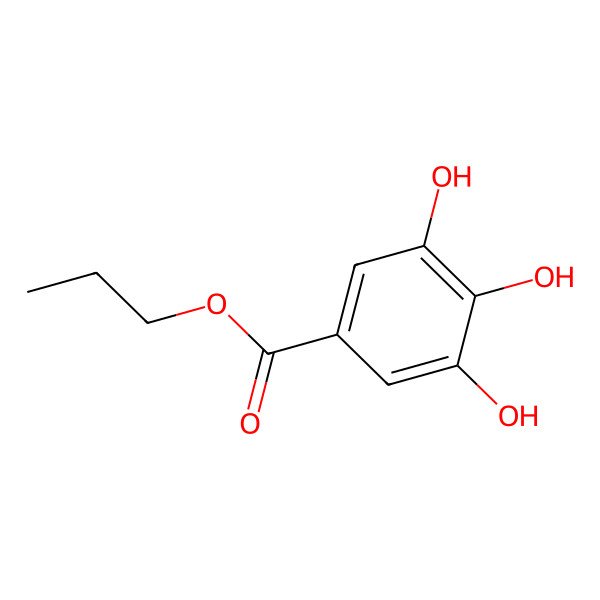
| Propyl 3,4,5-trihydroxybenzoate |
| N-Propyl gallate |
| Tenox PG |
| Progallin P |
| Gallic acid, propyl ester |
| Nipagallin P |
| Gallic acid propyl ester |
| NIPA 49 |
| 3,4,5-Trihydroxybenzoic acid propyl ester |
| Benzoic acid, 3,4,5-trihydroxy-, propyl ester |
| n-Propyl 3,4,5-trihydroxybenzoate |
| FEMA No. 2947 |
| 3,4,5-Trihydroxybenzene-1-propylcarboxylate |
| Propylester kyseliny gallove |
| n-Propyl ester of 3,4,5-trihydroxybenzoic acid |
| Pro gallin P |
| Gallic acid n-propyl ester |
| NSC 2626 |
| 3,4,5-Trihydroxybenzoic acid, propyl ester |
| NCI-C505888 |
| 3,4,5-Trihydroxybenzoic acid n-propyl ester |
| CCRIS 541 |
| Nipanox S 1 |
| HSDB 591 |
| Propyl gallate (NF) |
| Propyl gallate [NF] |
| C10H12O5 |
| NSC-2626 |
| EINECS 204-498-2 |
| Propylester kyseliny gallove [Czech] |
| Propyl gallate (e 310) |
| CHEMBL7983 |
| Gallic acid, n-propyl ester |
| UNII-8D4SNN7V92 |
| E310 |
| AI3-17136 |
| 8D4SNN7V92 |
| INS NO.310 |
| DTXSID5021201 |
| CHEBI:10607 |
| INS-310 |
| NSC2626 |
| Anhydrous propyl gallate (e 310) |
| NCGC00164234-01 |
| Gallic acid-propyl ester |
| E-310 |
| DTXCID901201 |
| Gallate, Propyl |
| CAS-121-79-9 |
| n-Propyl-3,4,5-Trihydroxybenzoate |
| Propyl galiate |
| l ester |
| n-propyl-gallate |
| Sustane PG |
| Propylgallate,(S) |
| Propyl Gallate FCC |
| MFCD00002196 |
| Propyl gallate, powder |
| Propyl gallate, 98% |
| 3,4,5-Trihydroxy-benzoic acid propyl ester |
| GALLATE, N-PROPYL |
| Gallic acid propyl esterZ |
| PROPYL GALLATE [II] |
| PROPYL GALLATE [MI] |
| Oprea1_580415 |
| SCHEMBL22630 |
| CBDivE_013134 |
| PROPYL GALLATE [FCC] |
| BIDD:ER0334 |
| Propyl 3,5-trihydroxybenzoate |
| PROPYL GALLATE [FHFI] |
| PROPYL GALLATE [HSDB] |
| PROPYL GALLATE [INCI] |
| WLN: QR BQ CQ EVO3 |
| PROPYL GALLATE [VANDF] |
| PROPYL GALLATE [MART.] |
| PROPYL GALLATE [USP-RS] |
| PROPYL GALLATE [WHO-DD] |
| FEMA 2947 |
| n-Propyl 3,5-trihydroxybenzoate |
| Propyl gallate, >=98%, FCC |
| NCI-C50588 |
| BCP13340 |
| HY-N0524 |
| propyl 3,4,5 - trihydroxybenzoate |
| Tox21_113531 |
| Tox21_202286 |
| Tox21_300060 |
| BDBM50032154 |
| LS-655 |
| s5113 |
| 3,4,5-Trihydroxy-benzoic acid propy |
| Propyl 3, 4, 5- trihydroxybenzoate |
| PROPYL GALLATE [EP MONOGRAPH] |
| AKOS001603853 |
| 5-Methyl-4,5-dihydrothiazole-2-thiol |
| CCG-207932 |
| DB12450 |
| NCGC00164234-02 |
| NCGC00164234-03 |
| NCGC00164234-04 |
| NCGC00254138-01 |
| NCGC00259835-01 |
| 3,5-Trihydroxybenzene-1-propylcarboxylate |
| 3,5-Trihydroxybenzoic acid, propyl ester |
| AC-11365 |
| AC-34485 |
| AS-11986 |
| NCI60_002094 |
| Propyl gallate, USP, 98.0-102.0% |
| Benzoic acid,4,5-trihydroxy-, propyl ester |
| CS-0009059 |
| EU-0036319 |
| FT-0626599 |
| G0018 |
| BENZOATE, 3,4,5-TRIHYDROXY-, PROPYL |
| n-Propyl ester of 3,5-trihydroxybenzoic acid |
| A19435 |
| D02382 |
| E80666 |
| Propyl gallate, antioxidant, >=98.0% (HPLC) |
| Q608726 |
| SR-01000944710 |
| Propyl gallate, for microscopy, >=98.0% (HPLC) |
| Q-201634 |
| SR-01000944710-1 |
| EFFF5FFA-651C-4DE3-A25F-D807C65D5537 |
| N-PROPYL ESTER OF 3,4,5- TRIHYDROXYBENZOIC ACID |
| Propyl gallate, European Pharmacopoeia (EP) Reference Standard |
| Propyl gallate, United States Pharmacopeia (USP) Reference Standard |
| Propyl gallate, Pharmaceutical Secondary Standard; Certified Reference Material |
| InChI=1/C10H12O5/c1-2-3-15-10(14)6-4-7(11)9(13)8(12)5-6/h4-5,11-13H,2-3H2,1H |
|
There are more than 10 synonyms. If you wish to see them all click here.
|