| JASMONE |
| 488-10-8 |
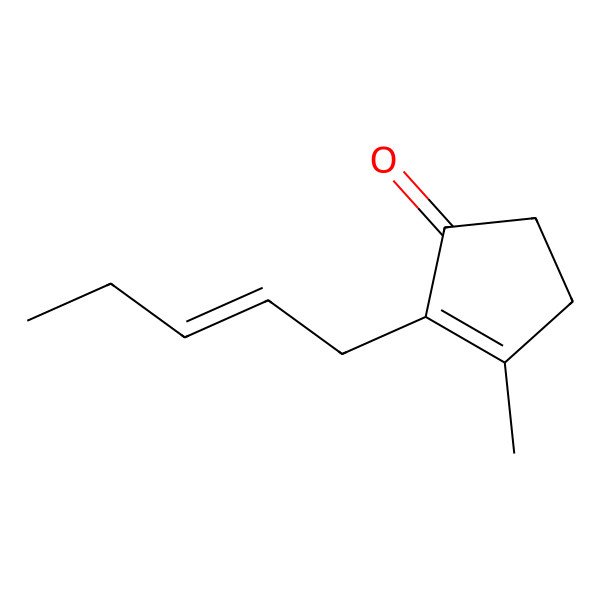
| (Z)-Jasmone |
| cis-?Jasmone |
| FEMA No. 3196 |
| cis-3-Methyl-2-(2-pentenyl)-2-cyclopenten-1-one |
| (Z)-3-Methyl-2-(pent-2-en-1-yl)cyclopent-2-enone |
| 2-Cyclopenten-1-one, 3-methyl-2-(2-pentenyl)-, (Z)- |
| 3-Methyl-2-pent-2-enylcyclopent-2-enone |
| 3-Methyl-2-(cis-2-penten-1-yl)-2-cyclopenten-1-one |
| EINECS 207-668-4 |
| UNII-RC4W0G9YUK |
| RC4W0G9YUK |
| BRN 1907713 |
| 3-methyl-2-[(Z)-pent-2-enyl]cyclopent-2-en-1-one |
| 3-Methyl-2-(2-pentenyl)-2-cyclopenten-1-one, (Z)- |
| CHEBI:6084 |
| DTXSID4047092 |
| 3-Methyl-2-(2-pentenyl)-2-cyclopenten-1-one |
| 3-methyl-2-(pent-2Z-enyl)cyclopent-2-enone |
| 3-Methyl-2-n-penten-2'-ylcyclopenten-2-one |
| 3-Methyl-2-(2-cis-pentenyl)-2-cyclopenten-1-one |
| 3-Methyl-2-(cis-2-pentenyl)-2-cyclopenten-1-one |
| 4-07-00-00337 (Beilstein Handbook Reference) |
| 2-Cyclopenten-1-one, 3-methyl-2-(2Z)-2-penten-1-yl- |
| 3-methyl-2-[(2Z)-pent-2-en-1-yl]cyclopent-2-en-1-one |
| 2-Cyclopenten-1-one, 3-methyl-2-(2Z)-2-pentenyl- (9CI) |
| 2-Cyclopenten-1-one, 3-methyl-2-(2-pentenyl)-, (Z)- (8CI) |
| MFCD00001402 |
| JASMONE, CIS- |
| cis-Jasmon |
| jasmone, cis |
| Jasmone (6CI) |
| Jasmone, 90% |
| cis-3-methyl-2-pent-2-enyl-cyclopent-2-enone |
| cis-Jasmone, stabilized |
| JASMONE [FHFI] |
| JASMONE [MI] |
| 2-ciclopenten-1-ona, 3-metil-2-(2z)-2-penten-1-il- |
| 3-Methyl-2-(2Z)-2-penten-1-yl-2-cyclopenten-1-one |
| Jasmone, analytical standard |
| 3 - methyl - 2 - pent - 2 - enylcyclopent - 2 - enone |
| SCHEMBL20385 |
| CHEMBL2251602 |
| DTXCID2027092 |
| HSDB 8273 |
| BCP18948 |
| CMC_7381 |
| HY-N7058 |
| Tox21_301825 |
| LMFA02020009 |
| s4960 |
| AKOS024318891 |
| CCG-266315 |
| CS-W017899 |
| GS-3251 |
| LS-2951 |
| NCGC00256139-01 |
| CAS-488-10-8 |
| J0003 |
| C08490 |
| EN300-7400412 |
| Q418077 |
| (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one |
| 3-Methyl-2-[(2Z)-2-pentenyl]-2-cyclopenten-1-one |
| 3-methyl-2-[(Z)-pent-2-enyl]-cyclopent-2-en-1-one |
| 2-Cyclopenten-1-one, 3-methyl-2-(2Z)-2- pentenyl- |
| 2-cyclopenten-1-one, 3-methyl-2-[(2Z)-2-pentenyl]- |
| 3-Methyl-2-[(2Z)-2-pentenyl]-2-cyclopenten-1-one # |
| METHYL-2-(2-PENTENYL)-2-CYCLOPENTEN-1-ONE, (Z)-, 3- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|