| 434-16-2 |
| Provitamin D3 |
| Dehydrocholesterol |
| 7,8-Didehydrocholesterol |
| Cholesta-5,7-dien-3beta-ol |
| 7-DHC |
| (-)-7-Dehydrocholesterol |
| Dehydrocholesterin |
| (3beta)-Cholesta-5,7-dien-3-ol |
| 7-Dehydrocholesterin |
| delta7-Cholesterol |
| Cholesta-5,7-dien-3-ol |
| 5,7-Cholestadien-3-beta-ol |
| delta5,7-Cholesterol |
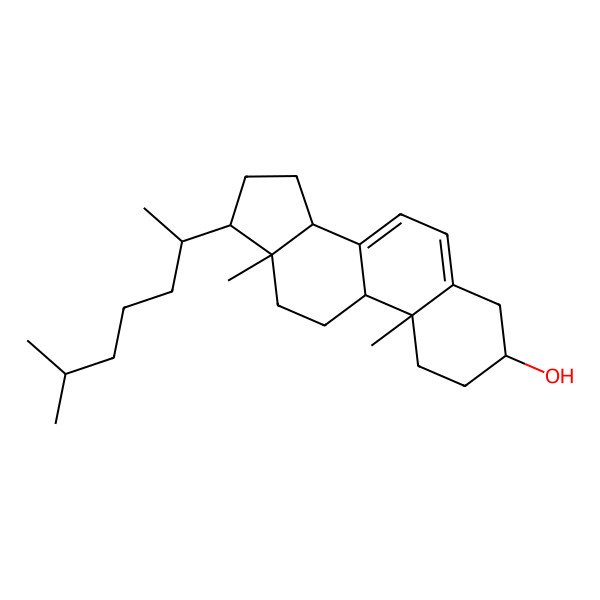
| (3S,9S,10R,13R,14R,17R)-10,13-dimethyl-17-[(2R)-6-methylheptan-2-yl]-2,3,4,9,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-ol |
| NSC 18159 |
| 5,7-Cholestandien-3beta-ol |
| (3beta)-7-Dehydro Cholesterol |
| 7-dehydro-cholesterol |
| Provitamin D(sub 3) |
| Caswell No. 902E |
| UNII-BK1IU07GKF |
| Cholesta-5,7-dien-3-ol, (3b)- |
| delta(sup 7)-Cholesterol |
| BK1IU07GKF |
| Dehydrocholesterin [German] |
| delta(sup 5,7)-Cholesterol |
| Cholesterol, 7-dehydro- |
| (3S,9S,10R,13R,14R,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,9,10,11,12,13,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
| CHEBI:17759 |
| EINECS 207-100-5 |
| EPA Pesticide Chemical Code 208700 |
| (3-BETA)-CHOLESTA-5,7-DIEN-3-OL |
| EC 207-100-5 |
| Cholesta-5,7-dien-3-beta-ol |
| Cholesta-5,7-dien-3.beta.-ol |
| Cholesta-5,7-dien-3beta-ol (7,8-Didehydrocholesterol; Provitamin D3) |
| Cholesta-5,7-dien-3-ol, (3.beta.)- |
| Cholesta-5,7-dien-3-ol # |
| 5,7-Cholestadien-3-.beta.-ol |
| 5,7-cholestadien-3beta-ol |
| cholesta-5,7-dien-3 beta-ol |
| NSC-18159 |
| provitamin-D3 |
| Cholesta-5,7-dien-3-ol, (3beta)- |
| .DELTA.5,7-Cholestadien-3.beta.-ol |
| Cholesta-5,7-dien-3-ol, (3-beta)- |
| MFCD00003624 |
| ?7-Dehydrocholesterol |
| 7,8-dehydro-cholesterol |
| 7DHC |
| 5,7-Cholestandien-3-ol |
| cholesta-5,7-dien-3b-ol |
| SCHEMBL3447 |
| 7, 8- didehydrocholesterol |
| 5,7-Cholestadien-3-bet-ol |
| (3ss)-7-Dehydro Cholesterol |
| (3-beta)-7-Dehydrocholesterol |
| CHEMBL1797131 |
| cholesta-5,7-dien-3 beta -ol |
| Delta5,7-Cholestadien-3beta-ol |
| DTXSID20861933 |
| (3b)-Cholesta-5,7-dien-3-ol |
| 3-bet.-Hydroxy-5,7-cholestadiene |
| 7-DEHYDROCHOLESTEROL [INCI] |
| LMST01010069 |
| s5386 |
| 7-Dehydrocholesterol (Provitamin D3) |
| 5,7-Cholestadien-3beta-ol monohydrate |
| CCG-268465 |
| CS-I-00099 |
| CS-T-51713 |
| AS-59257 |
| 7-Dehydrocholesterol, >=95.0% (HPLC) |
| HY-113279 |
| CS-0059496 |
| (3.BETA.)-7-DEHYDROCHOLESTEROL [MI] |
| C01164 |
| F20429 |
| CHOLECALCIFEROL IMPURITY B [EP IMPURITY] |
| Q139350 |
| W-106230 |
| 9.BETA.,10.ALPHA.-CHOLESTA-5,7-DIEN-3.BETA.-OL |
| 10,13-dimethyl-17-(6-methylheptan-2-yl)-2,3,4,9,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-ol |
| 17-(1,5-dimethylhexyl)-10,13-dimethyl-2,3,4,9,10,11,12,13,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
|
There are more than 10 synonyms. If you wish to see them all click here.
|