| 1463-10-1 |
| Ribothymidine |
| Thymine riboside |
| Ribosylthymine |
| Uridine, 5-methyl- |
| ribosylthymidine |
| beta-D-Ribofuranoside |
| Thymine ribofuranoside |
| 1-(beta-D-ribofuranosyl)thymine |
| UNII-ZS1409014A |
| CHEBI:45996 |
| EINECS 215-973-9 |
| ZS1409014A |
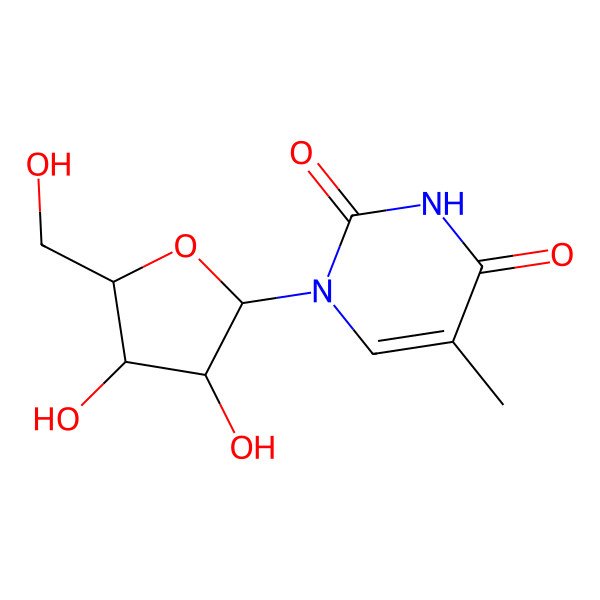
| 1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione |
| 1-((2R,3R,4S,5R)-TETRAHYDRO-3,4-DIHYDROXY-5-(HYDROXYMETHYL)FURAN-2-YL)-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE |
| C10H14N2O6 |
| MFCD00006535 |
| 5-methyl-Uridine |
| 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2,3,4-tetrahydropyrimidine-2,4-dione |
| 1-[(4S,2R,3R,5R)-3,4-Dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,3-dihydropyrimidine-2,4-dione |
| 5MU-5MU-5MU |
| Thymine ribonucleoside |
| 5-Methyluridine, powder |
| beta-delta-Ribofuranoside |
| 5-Methyluridine, 97% |
| bmse000759 |
| Epitope ID:141497 |
| 1-b-D-Ribofuranosylthymine |
| SCHEMBL29298 |
| b-D-Ribofuranoside thymine-1 |
| .beta.-D-Ribofuranosylthymine |
| THYMINE RIBOSIDE [MI] |
| CHEMBL106175 |
| 1-.beta.-D-Ribofuranosylthymine |
| CHEBI:30821 |
| 1-beta-delta-Ribofuranosylthymine |
| DTXSID20163348 |
| THYMINE RIBOSIDE, (-)- |
| beta-delta-Ribofuranoside thymine-1 |
| .beta.-D-Ribofuranoside, thymine-1 |
| s6156 |
| STL507242 |
| thymine-1 beta-D-Ribofuranosylthymine |
| AKOS015896830 |
| AC-8172 |
| AM83947 |
| CS-W010160 |
| HY-W009444 |
| 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidine-2,4-dione |
| thymine-1 beta-delta-Ribofuranosylthymine |
| BP-58644 |
| DS-18476 |
| PD099507 |
| M1405 |
| EN300-2009864 |
| A808481 |
| Q425078 |
| J-700101 |
| Z3037473449 |
| 5-methyl-1-beta-D-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione |
| PYRIMIDINEDIONE, 5-METHYL-1-.BETA.-D-RIBOFURANOSYL- |
| 2,4(1H,3H)-Pyrimidinedione, 5-methyl-1-.beta.-D-ribofuranosyl- |
| 5-methyl-1-beta-delta-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione |
| 1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)-tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione |
| 1-.BETA.-D-RIBOFURANOSYLTHYMINE 2,4(1H,3H)-PYRIMIDINEDIONE, 5-METHYL-1-.BETA.-D-RIBOFURANOSYL- |
| 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-5-methyl-pyrimidine-2,4-dione |
| 38T |
|
There are more than 10 synonyms. If you wish to see them all click here.
|