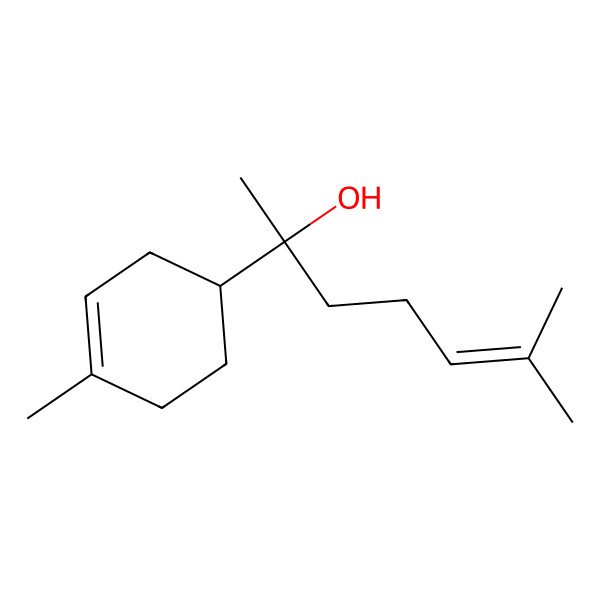
| (-)-alpha-Bisabolol |
| 23089-26-1 |
| Kamillosan |
| alpha-Bisabolol |
| Bisabolol |
| alpha-(-)-Bisabolol |
| Levomenolum |
| Kamilosan |
| .alpha.-Bisabolol |
| 1-alpha-Bisabolol |
| Levomenol [INN] |
| alpha-Bisabolol, L- |
| alpha-Bisabolol (-)-form |
| UNII-24WE03BX2T |
| 24WE03BX2T |
| CCRIS 9081 |
| (-)-6-Methyl-2-(4-methyl-3-cyclohexen-1-yl)-5-hepten-2-ol |
| DTXSID4042094 |
| FEMA NO. 4666 |
| (2S)-6-methyl-2-[(1S)-4-methylcyclohex-3-en-1-yl]hept-5-en-2-ol |
| (-)-.alpha.-Bisabolol |
| EINECS 245-423-3 |
| Levomenolum [INN-Latin] |
| (-)--BISABOLOL |
| Bisabolol, (-)-.alpha. |
| .alpha.-bisabolol, (-)- |
| CHEBI:125 |
| .alpha.-Bisabolol (-)-form |
| DTXCID2022094 |
| (.alpha.s,1s)-.alpha.-bisabolol |
| 3-Cyclohexene-1-methanol, alpha,4-dimethyl-alpha-(4-methyl-3-pentenyl)-, (alphaS,1S)- |
| LEVOMENOL (MART.) |
| LEVOMENOL [MART.] |
| 3-Cyclohexene-1-methanol, a,4-dimethyl-a-(4-methyl-3-pentenyl)-,(aS,1S)- |
| LEVOMENOL (USP-RS) |
| LEVOMENOL [USP-RS] |
| Dragosantol |
| Camilol |
| a-Bisabolol |
| Hydagen B |
| ((-))-6-Methyl-2-(4-methyl-3-cyclohexen-1-yl)-5-hepten-2-ol |
| (S)-6-Methyl-2-((S)-4-methylcyclohex-3-en-1-yl)hept-5-en-2-ol |
| 5-Hepten-2-ol, 6-methyl-2-(4-methyl-3-cyclohexen-1-yl)-, (-)- |
| 3-Cyclohexene-1-methanol, .alpha.,4-dimethyl-.alpha.-(4-methyl-3-pentenyl)-, [S-(R*,R*)]- |
| 3-Cyclohexene-1-methanol, alpha,4-dimethyl-alpha-(4-methyl-3-pentenyl)-, (S-(theta,theta))- |
| BISABOLOL, ALPHA |
| BISABOLOL, .ALPHA. |
| RACEMIC ALFA-BISABOLOL |
| 515-69-5 |
| Bisbalol |
| BISABOLA-1,12-DIEN-8-OL |
| EINECS 208-205-9 |
| EINECS 246-973-7 |
| L-alpha-Bisabolol |
| Levomenol, INN |
| (-)-a-Bisabolol |
| l-.alpha.-Bisabolol |
| .alpha.-Bisabolol, L- |
| BISABOLOL [INCI] |
| (-)- alpha -Bisabolol |
| .alpha.-(-)-Bisabolol |
| BISABOLOL [VANDF] |
| ALPHA-(-)BISABOLOL |
| LEVOMENOL [WHO-DD] |
| BISABOLOL, (-)-alpha |
| SCHEMBL24989 |
| alpha-BISABOLOL, (-)- |
| (alphaS,1S)-alpha-BISABOLOL |
| 3-Cyclohexene-1-methanol, ?,4-dimethyl-?-(4-methyl-3-pentenyl)-, (R,R)- |
| CHEMBL1096927 |
| (-)-(4S,8S)-alpha-Bisabolol |
| (-)-alpha-Bisabolol (Levomenol) |
| (alpha R,1R)-rel-alpha,4-Dimethyl-alpha-(4-methyl-3-penten-1-yl)-3-cyclohexene-1-methanol |
| (R*, R*)- a, 4- dimethyl- a- (4- methyl- 3- pentenyl)cyclohex- 3- ene- 1- methanol |
| (R*,R*)-alpha,4-Dimethyl-alpha-(4-methyl-3-pentenyl)cyclohex-3-ene-1-methanol |
| (R*,R*) - a,4 - dimethyl - a - (4 - methyl - 3 - pentenyl)cyclohex - 3 - ene - 1 - methanol |
| 3-ciclohexeno-1-metanol, Alfa,4-Dimetil-Alfa-(4-metil-3-penten-1-il)-, (Alfa R, 1R)-rel- |
| HY-N6967 |
| [S-(R*,R*)]-.alpha.-Bisabolol |
| Tox21_301375 |
| BDBM50382730 |
| MFCD03412455 |
| NSC782530 |
| Levomenol 100 microg/mL in Methanol |
| DB13153 |
| LMPR0103060001 |
| NSC-782530 |
| (-)-alpha-Bisabolol, >=93% (GC) |
| (-)-Alpha-bisabolol(-)-alpha-bisabolol |
| NCGC00255987-01 |
| (-)-alpha-Bisabolol, analytical standard |
| .ALPHA.-BISABOLOL (-)-FORM [MI] |
| CAS-23089-26-1 |
| CS-0028206 |
| Alpha-Bisabolol 1000 microg/mL in Isopropanol |
| C09621 |
| EN300-19632355 |
| Q179896 |
| J-014978 |
| (-)-alpha-BISABOLOL (CONSTITUENT OF CHAMOMILE) |
| (-)-alpha-Bisabolol, primary pharmaceutical reference standard |
| Levomenol, United States Pharmacopeia (USP) Reference Standard |
| (-)-.ALPHA.-BISABOLOL (CONSTITUENT OF CHAMOMILE) [DSC] |
| 6-Methyl-2-(4-methyl-3-cyclohexen-1-yl)-5-hepten-2-ol, (-)- |
| 5-HEPTEN-2-OL, 6-METHYL-2-(4- METHYL-3-CYCLOHEXEN-1-YL), (-)- |
| 5-HEPTEN-2-OL, 6-METHYL-2-(4-METHYL-3-CYCLOHEXEN-1-YL), (-)- |
| [S-(R*,R*)]-.alpha.,4-Dimethyl-.alpha.-(4-methyl-3-pentenyl)cyclohex-3-ene-1-methanol |
| 3-ciclohexeno-1-metanol, ?,4-dimetil-?-(4-metil-3-penten-1-il)-, (?s,1s)- |
| 3-Cyclohexene-1-methanol, .alpha.,4-dimethyl-.alpha.-(4-methyl-3-pentenyl)-, (-)- |
| 3-Cyclohexene-1-methanol, ?,4-dimethyl-?-(4-methyl-3-pentenyl)-, [S-(R,R)]- |
| 3-Cyclohexene-1-methanol, alpha,4-dimethyl-alpha-(4-methyl-3-pentenyl)-, (.alpha.S,1S)- |
| 3-Cyclohexene-1-methanol, alpha,4-dimethyl-alpha-(4-methyl-3-pentenyl)-, (S-(R*,R*))- |
| alpha,4-Dimethyl-alpha-(4-methyl-3-penten-1-yl)-(alphaS,1S)-3-Cyclohexene-1-methanol |
| 3-CYCLOHEXENE-1-METHANOL, .ALPHA.,4-DIMETHYL-.ALPHA.-(4-METHYL-3-PENTEN-1-YL)-, (.ALPHA.S,1S)- |
| 3-CYCLOHEXENE-1-METHANOL, alpha,4-DIMETHYL-alpha-(4-METHYL-3-PENTEN-1-YL)-, (alphaS,1S)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|