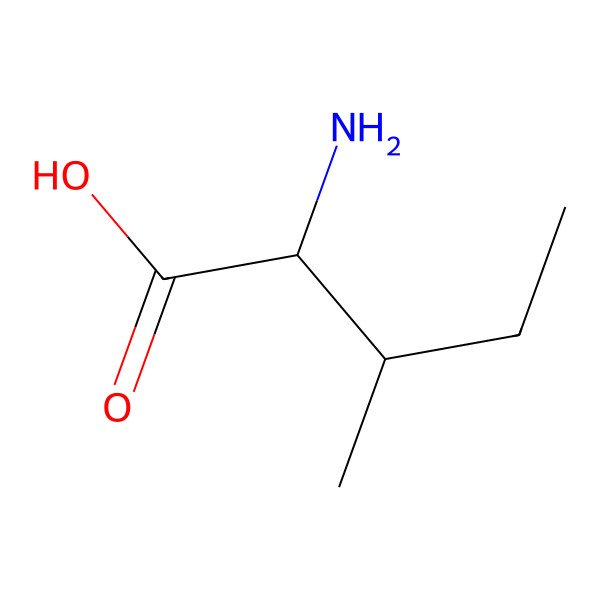
| Isoleucine |
| 73-32-5 |
| (2S,3S)-2-Amino-3-methylpentanoic acid |
| 2S,3S-Isoleucine |
| (S)-Isoleucine |
| H-Ile-OH |
| (S,S)-Isoleucine |
| 2-Amino-3-methylvaleric acid |
| L-(+)-Isoleucine |
| erythro-L-Isoleucine |
| L-Ile |
| ISOLEUCINE, L- |
| alpha-Amino-beta-methylvaleric acid |
| Isoleucine (VAN) |
| Isoleucinum [Latin] |
| Isoleucina [Spanish] |
| Norvaline, 3-methyl- |
| Valeric acid, 2-amino-3-methyl- |
| L-Norvaline, 3-methyl-, erythro- |
| Acetic acid, amino-sec-butyl- |
| ile |
| Pentanoic acid, 2-amino-3-methyl- |
| Isoleucina |
| Isoleucinum |
| CCRIS 5229 |
| iso-leucine |
| (2S,3S)-alpha-Amino-beta-methyl-n-valeric acid |
| Isoleucine [USAN:INN] |
| NSC 46708 |
| (2S,3S)-alpha-Amino-beta-merthylvaleric acid |
| (S-(R*,R*))-2-Amino-3-methylpentanoic acid |
| FEMA No. 3295 |
| (2S,3S)-alpha-Amino-beta-merthyl-n-valeric acid |
| 2-Amino-3-methylpentanoic acid, (S-(R*,R*))- |
| Acetic acid, amino(1-methylpropyl)-, (R*,R*)- |
| EINECS 200-798-2 |
| Isoleucine (L-Isoleucine) |
| UNII-5HX0BYT4E3 |
| 5HX0BYT4E3 |
| Pentanoic acid, 2-amino-3-methyl-, (S-(R*,R))- |
| (2S,3S)-2-Amino-3-methylpentanoicacid |
| DTXSID2046882 |
| FEMA NO. 4675 |
| CHEBI:17191 |
| HSDB 7798 |
| [S-(R*,R*)]-2-Amino-3-methylpentanoic acid |
| 2S-Amino-3S-methylpentanoic acid |
| Isoleucine, DL- |
| Isoleucine [USAN:USP:INN:BAN] |
| UNII-04Y7590D77 |
| DL-Allo-isoleucine |
| NSC 9958 |
| EINECS 207-139-8 |
| NSC-46708 |
| AI3-18474 |
| 04Y7590D77 |
| MFCD00064222 |
| (2S,3S)-alpha-Amino-beta-methylvaleric acid |
| Pentanoic acid, 2-amino-3-methyl-, (2S,3S)- |
| DTXCID201323037 |
| MFCD00004268 |
| DL- allo- isoleucine |
| CAS-443-79-8 |
| (2S,3S)-2-amino-3-methylpentanoate |
| Isoleucine, (2R,3R)-rel- |
| NSC-9958 |
| NSC46708 |
| L-isoleucina |
| laevo-isoleucine |
| l- isoleucine |
| L-iso-leucine |
| (L)-Isoleucine |
| L- iso-Leucine |
| NCGC00181164-01 |
| Isoleucine (USP) |
| H-Ile |
| Ile-OH |
| L-Isoleucine,(S) |
| (+)-L-isoleucine |
| L-[14C]Isoleucine |
| (2S,3S)-a-Amino-b-methylvaleric acid |
| (+/-)-erythro-2-Amino-3-methylpentanoic acid |
| L-Isoleucine, 99% |
| ISOLEUCINE [II] |
| ISOLEUCINE [MI] |
| (2S,3S)-a-Amino-b-methyl-n-valeric acid |
| L-Isoleucine (JP17) |
| ISOLEUCINE [INN] |
| (2S,3S)-2-amino-3-methyl-Pentanoic acid |
| ISOLEUCINE [HSDB] |
| ISOLEUCINE [INCI] |
| ISOLEUCINE [USAN] |
| ISOLEUCINE DL-FORM |
| 2-Amino-3-methylvalerate |
| ISOLEUCINE [VANDF] |
| Isoleucine, L- (8CI) |
| bmse000041 |
| bmse000866 |
| bmse000884 |
| D0I8SK |
| 2-amino-3-methylpentanoate |
| ISOLEUCINE [MART.] |
| L-Isoleucine (H-lle-OH) |
| L-ISOLEUCINE [FCC] |
| L-ISOLEUCINE [JAN] |
| SCHEMBL8869 |
| DL-ISOLEUCINE [FCC] |
| ISOLEUCINE [WHO-DD] |
| sec-C4H9CH(NH2)COOH |
| Acetic acid, amino-s-butyl- |
| DL-ISOLEUCINE [FHFI] |
| erythro-3-methyl-L-norvaline |
| L-Isoleucine (H-L-Ile-OH) |
| L-ISOLEUCINE [USP-RS] |
| GTPL3311 |
| CHEMBL1233584 |
| DTXSID1047441 |
| ISOLEUCINE DL-FORM [MI] |
| L-Isoleucine: D-allo-isoleucine |
| BDBM18140 |
| Norvaline, 3-methyl-, erythro- |
| ISOLEUCINE [EP MONOGRAPH] |
| ISOLEUCINE [USP MONOGRAPH] |
| L-Isoleucine, 99%, FCC, FG |
| Pharmakon1600-01301004 |
| (2S,3S)-a-Amino-b-methylvalerate |
| HY-N0771 |
| Tox21_112765 |
| LMFA01100047 |
| NSC760109 |
| s3752 |
| L-Isoleucine, Vetec(TM), 98.5% |
| AKOS015842027 |
| Tox21_112765_1 |
| (2S,3S)-a-Amino-b-methyl-n-valerate |
| AM81842 |
| CCG-266114 |
| CS-W018502 |
| DB00167 |
| FD20022 |
| NSC-760109 |
| (2S,3S)-2-amino-3-methyl-Pentanoate |
| VALINE IMPURITY B [EP IMPURITY] |
| (2S,3S)-alph-Amino-beta-methylvalerate |
| LEUCINE IMPURITY A [EP IMPURITY] |
| NCGC00274084-01 |
| (2S,3S)-alpha-Amino-beta-methylvalerate |
| AC-34995 |
| AS-11616 |
| BP-20357 |
| LS-84762 |
| (2S,3S)-alpha-Amino-beta-merthylvalerate |
| [S-(R*,R*)]-2-Amino-3-methylpentanoate |
| L-Isoleucine, BioUltra, >=99.5% (NT) |
| (2S,3S)-alph-Amino-beta-methylvaleric acid |
| (2S,3S)-alpha-Amino-beta-methyl-n-valerate |
| I0181 |
| (2S,3S)-alpha-Amino-beta-merthyl-n-valerate |
| EN300-52623 |
| L-Isoleucine, SAJ special grade, >=99.0% |
| C00407 |
| D00065 |
| L-Isoleucine, reagent grade, >=98% (HPLC) |
| M03002 |
| L-Isoleucine, Vetec(TM) reagent grade, >=98% |
| L-Isoleucine, Cell Culture Reagent (H-L-Ile-OH) |
| Q484940 |
| Q-201311 |
| (2S,3S)-.alpha.-Amino-.beta.-methyl-n-valeric acid |
| .alpha.-Amino-.beta.-methylvaleric acid, (2S,3S)- |
| Pentanoic acid, 2-amino-3-methyl-, [S-(R*,R*)]- |
| F8880-9085 |
| Z756427442 |
| Isoleucine, European Pharmacopoeia (EP) Reference Standard |
| L-Isoleucine, certified reference material, TraceCERT(R) |
| E46116A2-987C-4709-9E80-A64DA838D5A1 |
| L-Isoleucine, United States Pharmacopeia (USP) Reference Standard |
| 6-?amino-?5-?(ethylamino)?-?1-?methyl-2,?4(1H,?3H)?-?Pyrimidinedione |
| L-Isoleucine, Pharmaceutical Secondary Standard; Certified Reference Material |
| (S,S)-2-amino-3-methyl-pentanoicacid;(s,s)-isoleucine;[S-(R*,R*)]-2-Amino-3-methylpentanoic acid |
| 1160211-67-5 |
| L-Isoleucine, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|