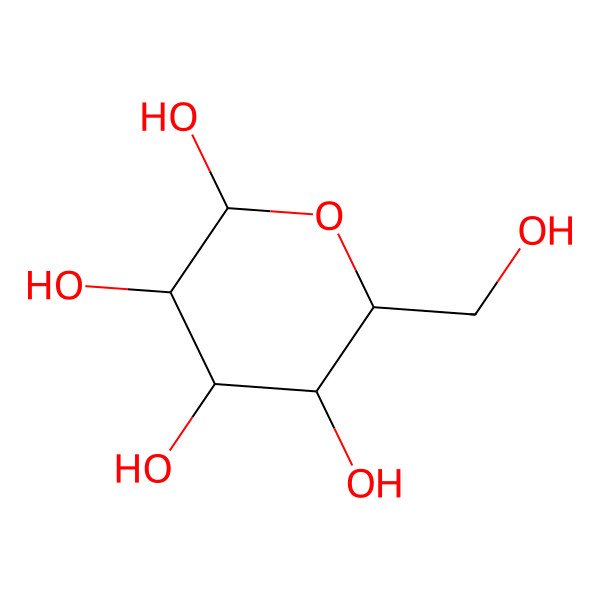
| alpha-D-Glucopyranose |
| 492-62-6 |
| alpha-glucose |
| alpha-Dextrose |
| Glucopyranose, alpha-D- |
| a-Dextrose |
| (2S,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol |
| a-D-Glucose |
| alpha-D-Glc |
| CHEBI:17925 |
| (2S,3R,4S,5S,6R)-6-(Hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetraol |
| glucoses |
| UNII-5J5I9EB41E |
| Grape sugar |
| 5J5I9EB41E |
| CHEMBL423707 |
| D-gluose |
| a-D-Glucopyranose |
| EINECS 207-757-8 |
| .alpha.-D-Glucopyranose |
| alpha-D-glucose, Phase I |
| alpha-D-glucose, Phase II |
| (1,4-alpha-D-Glucosyl)n |
| 27707-45-5 |
| alpha-D-glucopyranose, Phase I |
| D-gluco-hexose |
| alpha-D-glucopyranose, Phase II |
| .alpha.-D-Glucose |
| MFCD00063774 |
| (+)-Glucose |
| ?-D-Glucopyranose |
| D-Glucose-5-C-tD-glucose [MeSH: Glucose] |
| 26655-34-5 |
| DTXSID30197710 |
| a-Glucose |
| DextroseAnhydrate |
| (2RS,3R,4S,5S,6R)-6-(Hydroxymethyl)tetrahydropyran-2,3,4,5-tetrol |
| alpha-delta-Glucose |
| alpha-d(+)-glucose |
| I+/--D-Glucose |
| 1,3-alpha-D-Glucan |
| 1,4-alpha-D-Glucan |
| 1,6-alpha-D-Glucan |
| D-Glucose, .alpha.- |
| |A-D-Glucose anhydrous |
| alpha-delta-Glucopyranose |
| D(+)Glucose, Anhydrous |
| Dextrin from corn, p.a. |
| Glucose (alpha-D)-isomer |
| bmse000015 |
| bmse000791 |
| bmse000797 |
| bmse000855 |
| Epitope ID:144998 |
| MolMap_000023 |
| Alpha-D-Glucose, Anhydrous |
| SCHEMBL6222 |
| (1->3)-alpha-D-glucan |
| (1->4)-alpha-D-glucan |
| (1->6)-alpha-D-glucan |
| D-(+)-Glucose, anhydrous |
| (1,6-alpha-D-Glucosyl)n |
| MLS006011570 |
| Glucose, p.a., ACS reagent |
| (1->3)-alpha-D-glucopyranan |
| (1->4)-alpha-D-glucopyranan |
| (1,4-alpha-D-Glucosyl)n+1 |
| (1,4-alpha-D-Glucosyl)n-1 |
| [alpha-D-Glucosyl-(1,3)]n |
| alpha-D(+)-Glucose, anhydrous |
| CHEBI:15444 |
| CHEBI:18269 |
| CHEBI:18398 |
| CHEBI:28100 |
| CHEBI:28102 |
| D-(+)-Glucose, AR, anhydrous |
| WQZGKKKJIJFFOK-DVKNGEFBSA-N |
| [alpha-D-Glucosyl-(1,3)]n+1 |
| alpha-D-Glucose, anhydrous, 96% |
| CMC_6867 |
| Dextrin from corn, Type I, powder |
| D-(+) Glucose, analytical standard |
| D-(+)-Glucose, analytical standard |
| GLUCOSE, ANHYDROUS [WHO-IP] |
| (2S,3R,4S,5S,6R)-6-(hydroxymethyl)tetrahydropyran-2,3,4,5-tetrol |
| alpha-D-glucose; D-glucose; glucose |
| BDBM50351158 |
| s6028 |
| D-Glucose 1000 microg/mL in Water |
| AKOS015950677 |
| Glucose; (alpha-D-glucose; Dextrose) |
| D-(+)-Glucose, >=99% (GC) |
| D-Glucose 1000 microg/mL in Methanol |
| NCGC00160621-01 |
| NCGC00160621-03 |
| AC-15067 |
| BS-17112 |
| SMR004703328 |
| D-(+)-Glucose, >=99.5% (GC) |
| GLUCOSUM, ANHYDROUS [WHO-IP LATIN] |
| HY-128417 |
| CS-0099249 |
| D-(+)-Glucose, LR, anhydrous, >=99.5% |
| D-(+)-Glucose, tested according to Ph.Eur. |
| EN300-59169 |
| alpha-D-Glucose, SAJ first grade, >=98.0% |
| C00267 |
| D-(+)-Glucose, BioXtra, >=99.5% (GC) |
| D70945 |
| 4-{(1,4)-alpha-D-Glucosyl}(n-1)-D-glucose |
| alpha-D-Glucose, SAJ special grade, >=98.0% |
| D-Glucose (Dextrose), NIST(R) SRM(R) 917C |
| A871826 |
| W-204032 |
| D-(+)-Glucose, plant cell culture tested, BioReagent |
| D-(+)-Glucose, Vetec(TM) reagent grade, >=99.5% |
| Dextrin from corn, commercial grade, Type II, powder |
| Q23905965 |
| Dextrose, meets EP, BP, JP, USP testing specifications |
| Glucose, European Pharmacopoeia (EP) Reference Standard |
| Z905052654 |
| 074AD9E3-1FC7-485C-8A50-2B653D501E5B |
| D-(+)-Glucose, suitable for mouse embryo, >=99.5% (GC) |
| D-(+)-Glucose, 99.9 atom % 16O, 99.9 atom % 12C |
| Dextrose, United States Pharmacopeia (USP) Reference Standard |
| D-(+)-Glucose, anhydrous, free-flowing, Redi-Dri(TM), >=99.5% |
| Dextrose, meets EP, BP, JP, USP testing specifications, anhydrous |
| D-(+)-Glucose, BioUltra, anhydrous, >=99.5% (sum of enantiomers, HPLC) |
| D-(+)-Glucose, Hybri-Max(TM), powder, BioReagent, suitable for hybridoma |
| D-(+)-Glucose, meets analytical specification of Ph. Eur., BP, anhydrous |
| Dextrose, Pharmaceutical Secondary Standard; Certified Reference Material |
| D-(+)-Glucose, powder, BioReagent, suitable for cell culture, suitable for insect cell culture, suitable for plant cell culture, >=99.5% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|