| 29548-30-9 |
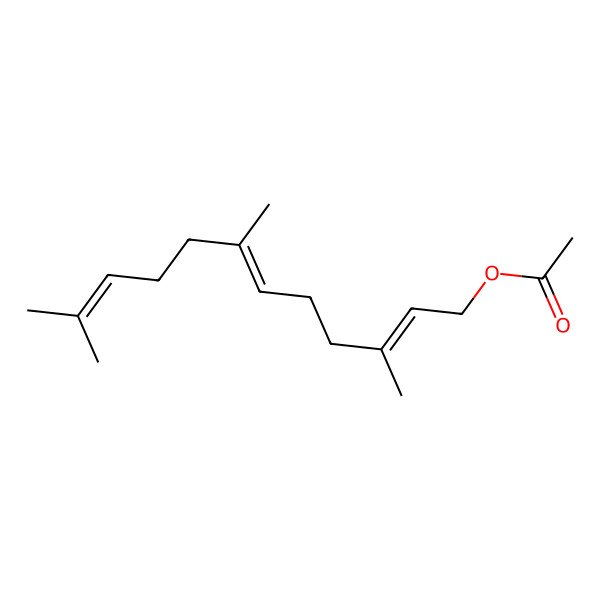
| trans,trans-Farnesyl acetate |
| 4128-17-0 |
| All-trans-Farnesyl acetate |
| (2E,6E)-Farnesyl acetate |
| 3,7,11-Trimethyl-2,6,10-dodecatrienyl acetate |
| [(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienyl] acetate |
| Farnesyl acetate, (2E,6E)- |
| (E,E)-farnesyl acetate |
| UNII-D5ZJ1FOC2I |
| D5ZJ1FOC2I |
| 2,6,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, acetate, (E,E)- |
| trans2,trans6-Farnesyl acetate |
| DTXSID5047110 |
| (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate |
| trans-2-trans-6-Farnesyl acetate |
| 2,6,10-dodecatrien-1-ol, 3,7,11-trimethyl-, acetate, (2E,6E)- |
| DTXSID2047222 |
| farneoylacetate |
| ((2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL) ACETATE |
| E,E-Farnesyl Acetate |
| (E)-Farnesyl acetate |
| 3,7,11 - trimethyldodeca - 2,6,10 - trienyl acetate |
| 2,6,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, 1-acetate |
| 3, 7, 11- trimethyldodeca- 2, 6, 10- trienyl acetate |
| Farnesyl acetate, (E,E)- |
| (cis,trans)-Farnesyl acetate |
| trans, trans-Farnesyl acetate |
| CHEMBL3184169 |
| DTXCID0027222 |
| DTXCID3027110 |
| farnesyl acetate, No Antioxidant |
| (2E,6E)-3,7,11-Trimethyl-2,6,10-dodecatrienyl acetate |
| 2-trans-6-trans-Farnesyl acetate |
| CHEBI:186251 |
| (E,E)-3,7,11-Trimethyl-2,6,10-dodecatrien-1-ol acetate |
| (E,E)-3,7,11-Trimethyl-2,6,10-dodecatrien-1-yl acetate |
| trans,trans-Farnesyl acetate, 95% |
| Tox21_302688 |
| Tox21_303761 |
| LMFA07010230 |
| MFCD00036516 |
| NSC132958 |
| s6150 |
| AKOS025295075 |
| AKOS037646546 |
| NSC-132958 |
| NCGC00256905-01 |
| NCGC00356962-01 |
| AS-69473 |
| CAS-4128-17-0 |
| CAS-29548-30-9 |
| HY-128430 |
| CS-0099265 |
| FEMA NO. 4213, (2E,6E)- |
| 3,11-Trimethyl-2,6,10-dodecatrienyl acetate |
| J-500791 |
| 2,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, acetate |
| 3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate |
| Q27276136 |
| (2E,6Z)-1-Acetoxy-3,7,11-trimethyl-2,6,10-dodecatriene |
| (2Z,6Z)-3,7,11-Trimethyl-2,6,10-dodecatriene-1-ol acetate |
| 2,6,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, acetate (E,E)- |
| Acetic acid (6E)-3,7,11-trimethyl-2,6,10-dodecatrienyl ester |
| [(E,E)-3,7,11-Trimethyl-2,6,10-dodecatriene-1-yl]ester of acetic acid |
| 2,6,10-DODECATRIEN-1-OL, 3,7,11-TRIMETHYL-, 1-ACETATE, (2E,6E)- |
| InChI=1/C17H28O2/c1-14(2)8-6-9-15(3)10-7-11-16(4)12-13-19-17(5)18/h8,10,12H,6-7,9,11,13H2,1-5H3/b15-10+,16-12 |
| Y7R |
|
There are more than 10 synonyms. If you wish to see them all click here.
|