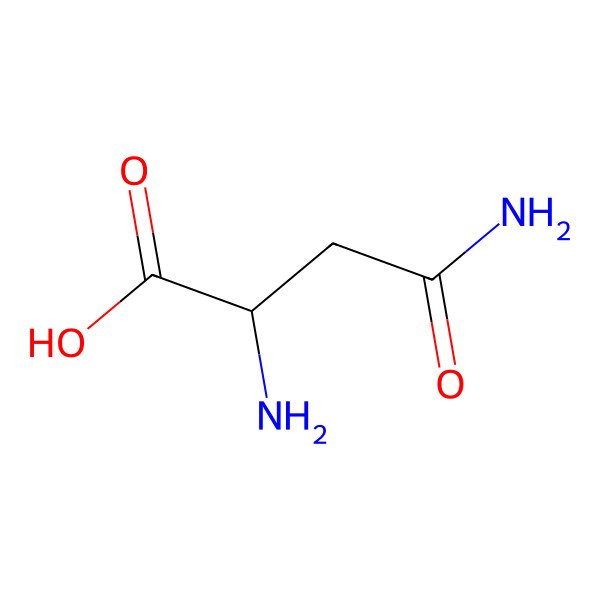
| asparagine |
| 70-47-3 |
| altheine |
| (S)-asparagine |
| Aspartamic acid |
| H-Asn-OH |
| agedoite |
| Asparamide |
| Crystal VI |
| asparagine acid |
| (S)-2,4-diamino-4-oxobutanoic acid |
| (-)-asparagine |
| 2-Aminosuccinamic acid |
| alpha-aminosuccinamic acid |
| Asparagine, L- |
| L-Asparagine anhydrous |
| L-Aspartamine |
| L-2,4-diamino-4-oxobutanoic acid |
| aspartic acid beta-amide |
| Asparagine anhydrous |
| ASN |
| L-asparatamine |
| (2S)-2-amino-3-carbamoylpropanoic acid |
| L-b-Asparagine |
| Asparagine (VAN) |
| 2-aminosuccinamic acid, L- |
| butanoic acid, 2,4-diamino-4-oxo-, (S)- |
| (2S)-2,4-diamino-4-oxobutanoic acid |
| L-2-aminosuccinamic acid |
| L-Asparagin |
| 7NG0A2TUHQ |
| L-beta-Asparagine (VAN) |
| L-aspartic acid beta-amide |
| L-beta-asparagine |
| NSC 82391 |
| L-Asn |
| 5Z33R5TKO7 |
| 2,4-diamino-4-oxobutanoic acid, (S)- |
| CHEBI:17196 |
| HSDB 7425 |
| EINECS 200-735-9 |
| MFCD00064401 |
| NSC-82391 |
| (S)-2-amino-3-carbamoylpropanoic acid |
| UNII-5Z33R5TKO7 |
| DTXSID10883220 |
| Aminoplasmal |
| Normofundin |
| Nutrifundin |
| Thomaeamin |
| (S)-2-Aminosuccinic acid 4-amide |
| asparagin |
| asparagina |
| L-Asparagin-1-wasser |
| Hasp |
| UNII-2PD79VF521 |
| ASPARAGINE ANHYDROUS (USP-RS) |
| ASPARAGINE ANHYDROUS [USP-RS] |
| L-(+)-Asparagine |
| L-.beta.-Asparagine |
| Aspartic acid b-amide |
| a-Aminosuccinamic acid |
| NSC 760099 |
| CHEBI:22653 |
| aspargine |
| Aspartamate |
| (-)-Asparagine; Asn; Asparamide |
| a-Aminosuccinamate |
| 2-Aminosuccinamate |
| Asn-OH |
| alpha-Aminosuccinamate |
| 4-imino-L-homoserine |
| alpha Amminosuccinamate |
| L-Asparagine (9CI) |
| H-Asn-OH H |
| ASPARAGINE [MI] |
| Aspartic acid beta amide |
| N-Lauryl-N-methyltaurine |
| ASPARAGINE [HSDB] |
| ASPARAGINE [INCI] |
| ASN NH3+ COOH |
| alpha Amminosuccinamic acid |
| Asparagine, L- (8CI) |
| bmse000030 |
| bmse000912 |
| Aspartic acid .beta. amide |
| L-ASPARAGINE [FCC] |
| L-Asparagine (H-Asn-OH) |
| ASPARAGINE [WHO-DD] |
| .alpha.-Aminosuccinamic acid |
| L-(+)-Asparagine anhydrous |
| CHEMBL58832 |
| ASPARAGINE (L) HYDRATE |
| Nor Benzphetamine Hydrochloride |
| (-)-Asparagine;Asn;Asparamide |
| GTPL4533 |
| p-aminosalicylicacidmagnesiumsalt |
| SCHEMBL3126352 |
| L-2,4-Diamino-4-oxobutanoate |
| US9138393, L-Asparigine |
| US9144538, L-Asparigine |
| BDBM181137 |
| DTXCID901022769 |
| Pharmakon1600-01301002 |
| (S)-2,4-Diamino-4-oxobutanoate |
| L-Asparagine, >=98% (HPLC) |
| HY-N0667 |
| STR07164 |
| B2,4-(S)-diamino-4-oxo-utanoate |
| NSC760099 |
| s5571 |
| AKOS006239067 |
| AC-4657 |
| AM81554 |
| CCG-266117 |
| DB00174 |
| NSC-760099 |
| B2,4-(S)-diamino-4-oxo-utanoic acid |
| NCGC00344576-01 |
| BP-23453 |
| A0542 |
| CS-0009702 |
| EN300-67209 |
| L-Asparagine, Vetec(TM) reagent grade, 98% |
| A-9031 |
| C00152 |
| M02998 |
| A937078 |
| Q29519883 |
| F1905-7151 |
| Z1066276474 |
| 3C28F6A9-E581-4255-ACCF-F75597AB288F |
| L-Asparagine, certified reference material, TraceCERT(R) |
| Asparagine anhydrous, United States Pharmacopeia (USP) Reference Standard |
| L-Asparagine, BioReagent, suitable for cell culture, suitable for insect cell culture |
|
There are more than 10 synonyms. If you wish to see them all click here.
|