| 141-22-0 |
| Ricinolic acid |
| Ricinic acid |
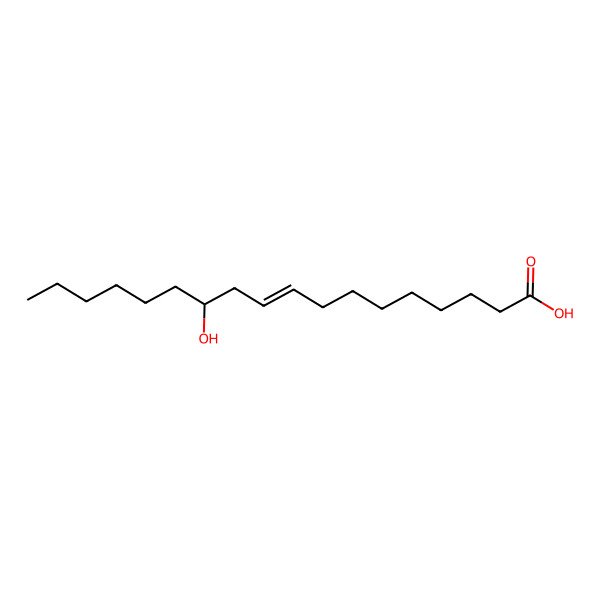
| (R,Z)-12-Hydroxyoctadec-9-enoic acid |
| Kyselina ricinolova |
| D-12-Hydroxyoleic acid |
| 12-Hydroxy-cis-9-octadecenoic acid |
| l'acide ricinoleique |
| (9Z,12R)-12-hydroxyoctadec-9-enoic acid |
| 12-hydroxyoleic acid |
| Acide ricinoleique |
| Oleic acid, 12-hydroxy- |
| Ricinolsaeure |
| Kyselina 12-hydroxy-9-oktadecenova |
| Nouracid CS 80 |
| NSC 281242 |
| 9-Octadecenoic acid, 12-hydroxy-, [R-(Z)]- |
| 12-Hydroxy-9-octadecenoic acid |
| UNII-I2D0F69854 |
| CHEBI:28592 |
| HSDB 5634 |
| EINECS 205-470-2 |
| 9-Octadecenoic acid, 12-hydroxy-, (9Z,12R)- |
| I2D0F69854 |
| (9Z,12R)-12-Hydroxy-9-octadecensaeure |
| (R-(Z))-12-Hydroxy-9-octadecenoic acid |
| 9-Octadecenoic acid, 12-hydroxy-, (Z)- |
| AI3-17956 |
| (9Z,12R)-12-Hydroxy-9-octadecenoic acid |
| 9-Octadecenoic acid, 12-hydroxy-, (cis)- |
| DTXSID0041567 |
| 12-Hydroxyoctadeca-9-enoic acid |
| 12R-hydroxy-9Z-octadecenoic acid |
| 9-Octadecenoic acid, 12-hydroxy-, (R-(Z))- |
| 9-Octadecenoic acid, 12-hydroxy- |
| (9Z)-(12R)-Hydroxyoctadecenoic acid |
| 12-D-Hydroxy-9-cis-octadecenoic acid |
| NSC-281242 |
| (Z,R)-12-hydroxyoctadec-9-enoic acid |
| 12-OH 9c-18:1 |
| (cis,R)-12-hydroxyoctadec-9-enoic acid |
| (R)-12-Hydroxy-cis-9-octadecenoic acid |
| 9-Octadecenoic acid, 12-hydroxy-, (theta-(Z))- |
| Riconoleic acid |
| DTXCID8021567 |
| RICINOLEIC ACID (MART.) |
| Ricinoleic acid (purity inverted exclamation markY99%) |
| RICINOLEIC ACID [MART.] |
| Ricinusoleic acid |
| [r]-12-hydroxy-cis-9-octadecenoic acid |
| CAS-141-22-0 |
| NCGC00161336-02 |
| Acide ricinoleique [French] |
| Kyselina ricinolova [Czech] |
| ricinoleic-acid |
| l'Acide ricinoleique [French] |
| 12-hydroxy-9-octadecenic acid |
| 9-Octadecenoic acid,12-hydroxy-,(Z)- |
| NCGC00181322-01 |
| MFCD00084840 |
| VESPULA PENSYLVANICA B708568K062 |
| (Z,12R)-12-hydroxyoctadec-9-enoic acid |
| UNII-KD8T3W6VZX |
| Kyselina 12-hydroxy-9-oktadecenova [Czech] |
| KD8T3W6VZX |
| Ricinoleic acid, >=99% |
| Ricinoleic acid, tech grade |
| SCHEMBL18144 |
| Flexricin 100 (Salt/Mix) |
| RICINOLEIC ACID [MI] |
| RICINOLEIC ACID [HSDB] |
| RICINOLEIC ACID [INCI] |
| BML2-F10 |
| RICINOLEIC ACID [VANDF] |
| 9-Octadecenoicacid,12-hydroxy- |
| CHEMBL3186422 |
| HY-N6070A |
| RICINOLEIC ACID [WHO-DD] |
| HY-N6070 |
| 12-Hydroxy-9(Z)-octadecenoic acid |
| cis-12-hydroxyoctadec-9-enoic acid |
| Ricinoleic acid, analytical standard |
| Tox21_113406 |
| Tox21_301098 |
| LMFA02000184 |
| 12R-Hydroxy-9-cis-octadecenoic Acid |
| AKOS016005826 |
| DB02955 |
| 9-Octadecenoic acid, 12-hydroxy-(cis) |
| NCGC00161336-01 |
| NCGC00161336-03 |
| NCGC00181089-01 |
| NCGC00254998-01 |
| (9Z)-12-Hydroxy-9-octadecenoic acid # |
| AS-65782 |
| CS-0032307 |
| CS-0204122 |
| R0027 |
| C08365 |
| F82175 |
| Ricinoleic acid, Vetec(TM) reagent grade, 80% |
| A885691 |
| EN300-21584706 |
| Q424484 |
| W-108189 |
| InChI=1/C18H34O3/c1-2-3-4-11-14-17(19)15-12-9-7-5-6-8-10-13-16-18(20)21/h9,12,17,19H,2-8,10-11,13-16H2,1H3,(H,20,21)/b12-9-/t17-/m1/s |
|
There are more than 10 synonyms. If you wish to see them all click here.
|