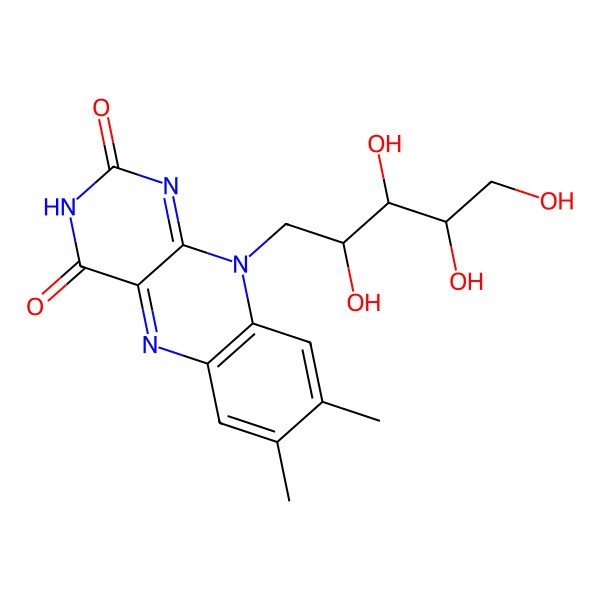
| vitamin B2 |
| 83-88-5 |
| Lactoflavin |
| Riboflavine |
| Vitamin G |
| Lactoflavine |
| Flavaxin |
| Riboflavinum |
| Beflavin |
| Beflavine |
| (-)-riboflavin |
| Lactobene |
| Ribocrisina |
| Riboderm |
| Riboflavina |
| Ribotone |
| Vitaflavine |
| Flaxain |
| Ribipca |
| Ribosyn |
| Ribovel |
| Vitamin Bi |
| Flavin BB |
| Russupteridine Yellow III |
| Vitasan B2 |
| Fiboflavin |
| Dermadram |
| Aqua-Flave |
| HYRE |
| Bisulase |
| Riboflavinequinone |
| 7,8-Dimethyl-10-ribitylisoalloxazine |
| 6,7-Dimethyl-9-D-ribitylisoalloxazine |
| E101 |
| Lacto-flavin |
| Hyflavin |
| vitamin-b2 |
| HSDB 817 |
| C.I. Food Yellow 15 |
| Riboflavine [INN-French] |
| Riboflavinum [INN-Latin] |
| Riboflavina [INN-Spanish] |
| Riboflavin (Vit B2) |
| CCRIS 1904 |
| Ins no.101(iii) |
| CHEBI:17015 |
| Ins-101(iii) |
| Riboflavin (Vitamin B2) |
| C17H20N4O6 |
| UNII-TLM2976OFR |
| 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)isoalloxazine |
| EINECS 201-507-1 |
| E-101(iii) |
| TLM2976OFR |
| Vitamin b2 (as riboflavin) |
| NSC 33298 |
| Vitamin B-2 |
| 7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]benzo[g]pteridine-2,4-dione |
| DTXSID8021777 |
| AI3-14697 |
| INS NO. 101(I) |
| Isoalloxazine, 7,8-dimethyl-10-D-ribityl- |
| C.I. 50900 |
| NSC-33298 |
| NCI-0033298 |
| Riboflavin [USP:INN:BAN] |
| Riboflavin [USAN:INN:JAN] |
| D-Ribitol, 1-deoxy-1-(3,4-dihydro-7,8-dimethyl-2,4-dioxobenzo(g)pteridin-10(2H)-yl)- |
| DTXCID401777 |
| Isoalloxazine, 7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)- |
| 1-Deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)pentitol |
| 7,8-dimethyl-10-((2S,3S,4R)-2,3,4,5-tetrahydroxypentyl)benzo[g]pteridine-2,4(3H,10H)-dione |
| Benzo(g)pteridine-2,4(3H,10H)-dione, 7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)- |
| RIBOFLAVIN (II) |
| RIBOFLAVIN [II] |
| MFCD00005022 |
| RIBOFLAVIN (MART.) |
| RIBOFLAVIN [MART.] |
| RIBOFLAVIN (USP-RS) |
| RIBOFLAVIN [USP-RS] |
| Vitamin B2 (Riboflavin) |
| 7,8-DIMETHYL-10-(1'-D-RIBITYL)ISOALLOXAZINE |
| RIBOFLAVIN (EP MONOGRAPH) |
| RIBOFLAVIN [EP MONOGRAPH] |
| RIBOFLAVIN (USP MONOGRAPH) |
| RIBOFLAVIN [USP MONOGRAPH] |
| 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-D-ribitol |
| 7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)benzo[g]pteridine-2,4(3H,10H)-dione |
| 7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]-2H,3H,4H,10H-benzo[g]pteridine-2,4-dione |
| Vitamin B 2 |
| 1-Deoxy-1-(3,4-dihydro-7,8-dimethyl-2,4-dioxobenzo[g]pteridin-10(2H)-yl)-D-ribitol |
| 1217461-14-7 |
| 7,8-dimethyl-10-ribityl-nu-isoalloxazine |
| vitaminum b2 |
| C17-H20-N4-O6 |
| 1kyv |
| 2ccb |
| 2vxa |
| ()-Riboflavin |
| 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo(g)pteridin-10(2H)-yl)-D-ribitol |
| 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo(g)pteridin-10(2H)-yl)pentitol |
| 5-deoxy-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo(g)pteridin-10(2H)-yl)-D-ribitol |
| 5-deoxy-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-D-ribitol |
| 7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)benzo(g)pteridine-2,4(3H,10H)-dione |
| San Yellow B |
| CAS-83-88-5 |
| NCGC00017291-05 |
| Bisulase (TN) |
| 7,8-dimethyl-10-((2S,3S,4R)-2,3,4,5-tetrahydroxypentyl)benzo(g)pteridine-2,4(3H,10H)-dione |
| Food Yellow 15 |
| Prestwick_442 |
| 2fl5 |
| 2vx9 |
| 4d1y |
| RIBOFLAVIN [MI] |
| RIBOFLAVIN [FCC] |
| RIBOFLAVIN [INN] |
| RIBOFLAVIN [JAN] |
| Vitamin B2; E101 |
| Prestwick3_000634 |
| RIBOFLAVIN [HSDB] |
| RIBOFLAVIN [INCI] |
| LACTOFLAVIN [INCI] |
| RIBOFLAVIN [VANDF] |
| D04QST |
| Epitope ID:161730 |
| RIBOFLAVINUM [HPUS] |
| SCHEMBL7706 |
| CHEMBL1534 |
| RIBOFLAVIN [WHO-DD] |
| RIBOFLAVIN [WHO-IP] |
| VITAMIN B2 [VANDF] |
| BSPBio_000628 |
| MLS001066391 |
| BPBio1_000692 |
| GTPL6578 |
| Riboflavin (JP17/USP/INN) |
| RIBOFLAVINE; VITAMIN B2 |
| RIBOFLAVIN [ORANGE BOOK] |
| A11HA04 |
| S01XA26 |
| VITAMIN B2 [GREEN BOOK] |
| (-)-Riboflavin, 97-103% |
| HMS2096P10 |
| Benzo[g]pteridine riboflavin deriv. |
| HY-B0456 |
| Riboflavin(B2);7,8-Dimethyl-10-((2S,3S,4R)-2,3,4,5-tetrahydroxypentyl)benzo[g]pteridine-2,4(3H,10H)-dione |
| RIBOFLAVINUM [WHO-IP LATIN] |
| Tox21_110813 |
| Tox21_111714 |
| Tox21_201633 |
| Tox21_302980 |
| 6,7-Dimethyl-9-ribitylisoalloxazine |
| BDBM50362895 |
| s2540 |
| Riboflavin (B2), analytical standard |
| Tox21_111714_1 |
| CS-O-31031 |
| DB00140 |
| LS-2192 |
| NCGC00091288-01 |
| NCGC00091288-02 |
| NCGC00091288-03 |
| NCGC00091288-04 |
| NCGC00091288-05 |
| NCGC00179498-01 |
| NCGC00256408-01 |
| NCGC00259182-01 |
| AS-15936 |
| SMR000112236 |
| R0020 |
| (-)-Riboflavin, tested according to Ph.Eur. |
| C00255 |
| D00050 |
| EN300-6477227 |
| (-)-Riboflavin, meets USP testing specifications |
| A840676 |
| A934900 |
| Q130365 |
| (-)-Riboflavin, from Eremothecium ashbyii, >=98% |
| W-104132 |
| Z2216887959 |
| RIBOFLAVIN SODIUM PHOSPHATE IMPURITY D [EP IMPURITY] |
| Riboflavin, European Pharmacopoeia (EP) Reference Standard |
| Riboflavin, United States Pharmacopeia (USP) Reference Standard |
| 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-isoalloxazine |
| Riboflavin, Pharmaceutical Secondary Standard; Certified Reference Material |
| 3,10-Dihydro-7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)benzopteridine-2,4-dione |
| 7,8-dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-Benzo[g]pteridine-2,4(3H,10H)-dione |
| d-ribitol, 1-Deoxy-1-(3,4-dihydro-7,8-dimethyl-2,4-dioxobenzo(g)pteridin-10(-2H)-yl)- |
| Riboflavin for peak identification, European Pharmacopoeia (EP) Reference Standard |
| (-)-Riboflavin, 100 mug/mL (1% ammonium acetate in 50:50 methanol:water), certified reference material, ampule of 1 mL |
| (-)-Riboflavin, BioReagent, suitable for cell culture, suitable for insect cell culture, >=98% |
| 7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrakis(oxidanyl)pentyl]benzo[g]pteridine-2,4-dione |
| 7,8-dimethyl-2,4-dioxo-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl](2,4a,10a-C,1,3-)-2H,3H,4H,10H-benzo[g]pteridin-3-yl |
| InChI=1/C17H20N4O6/c1-7-3-9-10(4-8(7)2)21(5-11(23)14(25)12(24)6-22)15-13(18-9)16(26)20-17(27)19-15/h3-4,11-12,14,22-25H,5-6H2,1-2H3,(H,20,26,27)/t11-,12+,14-/m0/s |
|
There are more than 10 synonyms. If you wish to see them all click here.
|