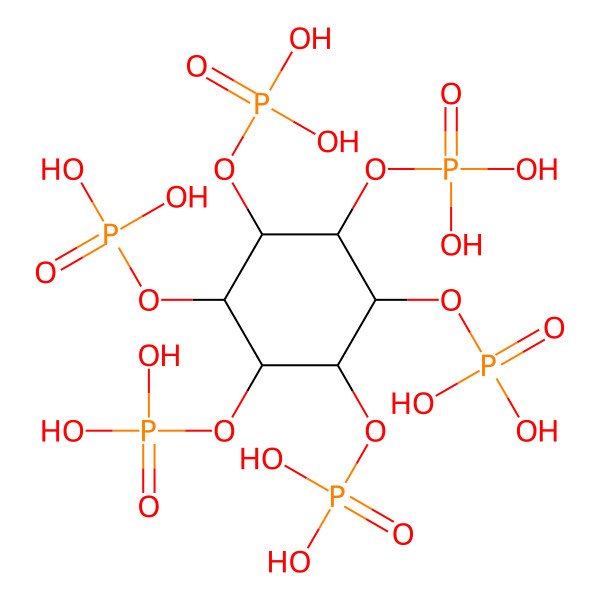
| 83-86-3 |
| Fytic acid |
| Phytate |
| Inositol hexaphosphate |
| Alkalovert |
| myo-inositol hexakisphosphate |
| myo-Inositol hexaphosphate |
| Alkovert |
| Phytine |
| Acide fytique |
| Acidum fyticum |
| Acido fitico |
| inositol hexakisphosphate |
| myo-Inositol, hexakis(dihydrogen phosphate) |
| Exfoderm |
| myo-Inosistol hexakisphosphate |
| (2,3,4,5,6-pentaphosphonooxycyclohexyl) dihydrogen phosphate |
| CCRIS 4513 |
| Phyliance |
| inositolhexaphosphoric acid |
| Dermofeel pa-3 |
| Saure des phytins |
| Inositol hexaphosphoric acid |
| Saure des phytins [German] |
| Saeure des phytins |
| Acide fytique [INN-French] |
| Acido fitico [INN-Spanish] |
| Acidum fyticum [INN-Latin] |
| Phytin |
| Fytic acid [INN] |
| Inosithexaphosphorsaure |
| Inosithexaphosphorsaure [German] |
| 1D-myo-inositol hexakisphosphate |
| Inositol 1,2,3,4,5,6-hexakisphosphate |
| CHEBI:17401 |
| Inosithexaphosphorsaeure |
| EINECS 201-506-6 |
| UNII-7IGF0S7R8I |
| NSC 269896 |
| NSC-269896 |
| 7IGF0S7R8I |
| INOSITOL, HEXAKIS(DIHYDROGEN PHOSPHATE), myo- |
| myo-inositol hexakis(dihydrogen phosphate) |
| IP6 |
| myo-Inositol, 1,2,3,4,5,6-hexakis(dihydrogen phosphate) |
| myo-Inositol 1,2,3,4,5,6-hexakisphosphate |
| D-chiro inositol hexakisphosphate |
| D-myo-Inositol 1,2,3,4,5,6-hexakisphosphate |
| 1D-myo-Inositol 1,2,3,4,5,6-hexakisphosphate |
| NSC269896 |
| Hexakis(dihydrogen phosphate) myo-inositol |
| Phytic acid (potassium) |
| Phyticacid |
| rel-(1R,2r,3S,4R,5s,6S)-Cyclohexane-1,2,3,4,5,6-hexayl hexakis(dihydrogen phosphate) |
| SNF-472 |
| IHP |
| meso-Inositol hexaphosphate |
| Inositol hexakis(phosphate) |
| fytinsyre- |
| myo-Inositol hexakis(phosphate) |
| hexasodium-phytate |
| 1zsh |
| (1R,2r,3S,4R,5s,6S)-cyclohexane-1,2,3,4,5,6-hexayl hexakis(dihydrogen phosphate) |
| C6H18O24P6 |
| Phytic Acid Powder |
| 1bq3 |
| neo-Inositol hexaphosphate |
| Phytic acid (dry powder) |
| PHYTIC ACID [MI] |
| D0F3GG |
| D0KN1U |
| Epitope ID:144992 |
| PHYTIC ACID [INCI] |
| scyllo-Inositol hexaphosphate |
| FYTIC ACID [MART.] |
| SCHEMBL19249 |
| FYTIC ACID [WHO-DD] |
| D-chiro-Inositol hexaphosphate |
| SCHEMBL136587 |
| GERD therapy (oral), Reviva |
| IP-6 |
| myo-inositol hexaphosphoric acid |
| SCHEMBL1681470 |
| SCHEMBL1681701 |
| SCHEMBL1682030 |
| SCHEMBL9164308 |
| SCHEMBL9741350 |
| SNF-03 |
| SNF-04 |
| CHEMBL1233511 |
| CHEMBL2005481 |
| SCHEMBL12137150 |
| SCHEMBL12138455 |
| SCHEMBL12549553 |
| SCHEMBL15374707 |
| SCHEMBL17006482 |
| diphosphoinositol tetrakisphosphate |
| DTXSID00861653 |
| DTXSID40889331 |
| SNF-471 |
| SNF-571 |
| SNF-671 |
| CHEBI:187038 |
| IMQLKJBTEOYOSI-GPIVLXJGSA-N |
| IMQLKJBTEOYOSI-OBXALCGXSA-N |
| IMQLKJBTEOYOSI-UHFFFAOYSA-N |
| HY-N0814 |
| MFCD00082309 |
| s3793 |
| AKOS015856604 |
| AKOS015901448 |
| C6-H18-O24-P6 |
| AC-8037 |
| CCG-270338 |
| CS-6330 |
| DB14981 |
| RP-3000 |
| NCGC00483012-01 |
| E391 |
| LS-84062 |
| NCI60_002200 |
| NCI60_038627 |
| PD006549 |
| PD008378 |
| PD054429 |
| Inositol Hexaphosporic Acid, Hexaphosphate |
| FT-0600486 |
| FT-0689105 |
| P0409 |
| D-myo-Inositol-1,2,3,4,5,6-hexaphosphate |
| A14529 |
| C01204 |
| A840671 |
| EN300-19650970 |
| Q409679 |
| SR-01000944523 |
| Phytic Acid (ca. 50% in Water, ca. 1.1mol/L) |
| Q-201578 |
| SR-01000944523-1 |
| Gastroesophageal reflux disease therapy (oral), Reviva |
| SNF-571 (transdermal patch, renal calcium lithiasis), Sanifit |
| {[2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
| cyclohexane-1,2,3,4,5,6-hexayl hexakis[dihydrogen (phosphate)] |
| Phytate (tablet formulation, osteoporosis), Sanifit Laboratoris SL |
| SNF-471 (transdermal patch, cardiovascular calcification), Sanifit |
| Phosphoric acid mono-(2,3,4,5,6-pentakis-phosphonooxy-cyclohexyl) ester |
| {[(1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
| InsP6 (transdermal patch formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| InsP6, (transdermal/patch formulation, renal calcium lithiasis), Sanifit Laboratoris SL |
| MYO-INOSITOL HEXAKISPHOSPHATE; INOSITOL 1,2,3,4,5,6-HEXAKISPHOSPHATE |
| Myo-inositol hexaphosphate (tablet formulation, osteoporosis), Sanifit Laboratoris SL |
| MYO-INOSITOL, 1,2,3,4,5,6-HEXAKIS(DIHYDROGEN PHOSPHATE),60% IN WATER |
| Phytate (intravenous formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Phytate (transdermal/patch formulation, renal calcium lithiasis), Sanifit Laboratoris SL |
| rel-(1R,2r,3S,4R,5s,6S)-Cyclohexane-1,2,3,4,5,6-hexaylhexakis(dihydrogenphosphate) |
| 1,2,3,4,5,6-cyclohexanehexol, hexakis(dihydrogen phosphate), (1alpha,2alpha,3alpha,4beta,5alpha,6beta)- |
| Calcium metabolism modulator (subcutaneous/injectable formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Calcium metabolism modulator (transdermal patch formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Myo-inositol hexakisphosphate (transdermal patchformulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Myo-inositol hexaphosphate (intravenous formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Myo-inositol hexaphosphate (transdermal patch formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
| Myo-inositol hexaphosphate (transdermal/patch formulation, renal calcium lithiasis), Sanfit Laboratoris SL |
| Phytate (transdermal patch formulation, cardiovascular calcifications), Sanifit Laboratoris SL |
|
There are more than 10 synonyms. If you wish to see them all click here.
|