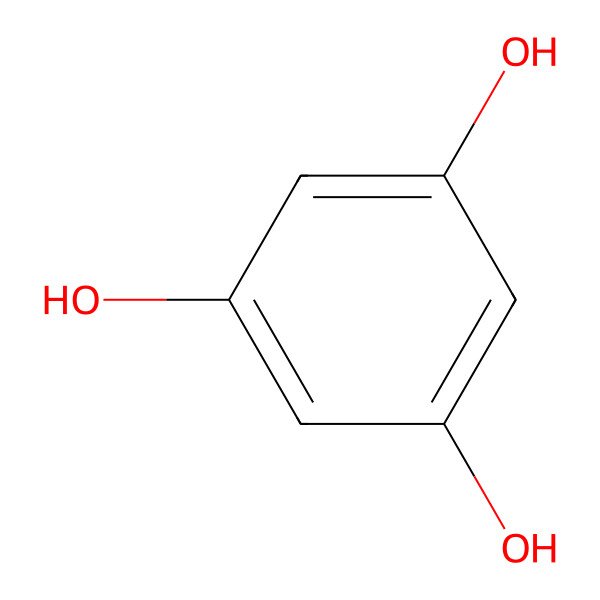
| 108-73-6 |
| Benzene-1,3,5-triol |
| 1,3,5-trihydroxybenzene |
| Phloroglucin |
| 1,3,5-benzenetriol |
| Phloroglucine |
| Spasfon-Lyoc |
| Dilospan S |
| sym-Trihydroxybenzene |
| s-Trihydroxybenzene |
| Benzene-s-triol |
| 5-Hydroxyresorcinol |
| Benzene, trihydroxy |
| 5-Oxyresorcinol |
| 3,5-Dihydroxyphenol |
| Floroglucin |
| Floroglucinol |
| Phloroglucinol anhydrous |
| 1,3,5-Triol |
| 1,3,5-THB |
| 5-Oxyresorcinolphloroglucin |
| 1,3,5-Trihydroxycyclohexatriene |
| Benzene, 1,3,5-trihydroxy- |
| Floroglucin [Czech] |
| NSC 1572 |
| Floroglucinol [Czech] |
| MFCD00002286 |
| CCRIS 4147 |
| DHD7FFG6YS |
| UNII-DHD7FFG6YS |
| Benezene-1,3,5-triol |
| 5-Benzenetriol |
| NSC-1572 |
| EINECS 203-611-2 |
| BRN 1341907 |
| Phloroglucinol (JAN) |
| CHEBI:16204 |
| AI3-08848 |
| Phloroglucinol-13C6 |
| CHEMBL473159 |
| 4-06-00-07361 (Beilstein Handbook Reference) |
| PHLOROGLUCINOL [JAN] |
| PHLOROGLUCINOL (MART.) |
| PHLOROGLUCINOL [MART.] |
| Phloroglucinol, anhydrous |
| PHLOROGLUCINOL (EP MONOGRAPH) |
| PHLOROGLUCINOL [EP MONOGRAPH] |
| NCGC00166270-01 |
| 1,2,5-trihydroxybenzene |
| benceno-s-triol |
| 1,5-Benzenetriol |
| 13X |
| Dilospan S (TN) |
| 3 5-Dihydroxyphenol |
| Phloroglucinol [BAN] |
| 1,3,5-Bencenotriol |
| 1,5-Trihydroxybenzene |
| 135-Trihydroxybenzene |
| Phloroglucinol (8CI) |
| 1,5-Triol |
| alpha-Aminoadipate, DL- |
| 1,3,5-trihidroxibenceno |
| Benzene,3,5-trihydroxy- |
| WLN: QR CQ EQ |
| 1,3,5-trihydroxylbenzene |
| D07EXH |
| 1,3,5-trihydroxy-benzene |
| PHLOROGLUCINOL [MI] |
| 1,3, 5-Trihydroxybenzene |
| (+-)-2-Aminoadipic acid |
| SCHEMBL26311 |
| PHLOROGLUCINOL [INCI] |
| 1,5-Trihydroxycyclohexatriene |
| 626-71-1 |
| Phloroglucinol, p.a., 99% |
| PHLOROGLUCINOL [WHO-DD] |
| 1,3,5-Benzenetriol, anhydrous |
| DTXSID9048354 |
| NSC1572 |
| BBL018701 |
| BDBM50249069 |
| STL146346 |
| phloroglucinol (1,3,5-benzenetriol) |
| Phloroglucinol, >=99.0% (HPLC) |
| AKOS000119851 |
| AM84333 |
| CS-T-39800 |
| DB12944 |
| FS-4574 |
| Hexanedioic acid, 2-amino-, (.+.)- |
| 1,3,5-Benzenetriol (ACD/Name 4.0) |
| NCGC00166270-02 |
| AC-10081 |
| LS-105905 |
| FT-0606511 |
| FT-0673853 |
| P0249 |
| P1376 |
| EN300-19648 |
| Phloroglucinol 100 microg/mL in Acetonitrile |
| C02183 |
| D00152 |
| F13526 |
| A801919 |
| Q899008 |
| Q-200070 |
| Phloroglucinol, plant cell culture tested, BioReagent |
| F0001-0177 |
| Z104474594 |
| Phloroglucinol, Used to detect the presence of wood fiber. |
| InChI=1/C6H6O3/c7-4-1-5(8)3-6(9)2-4/h1-3,7-9 |
| Phloroglucinol (anhydrous), European Pharmacopoeia (EP) Reference Standard |
|
There are more than 10 synonyms. If you wish to see them all click here.
|