Morin
| Internal ID | 3e422232-e80a-4ccd-b301-5929e2e6867f |
| Taxonomy | Phenylpropanoids and polyketides > Flavonoids > Flavones > Flavonols |
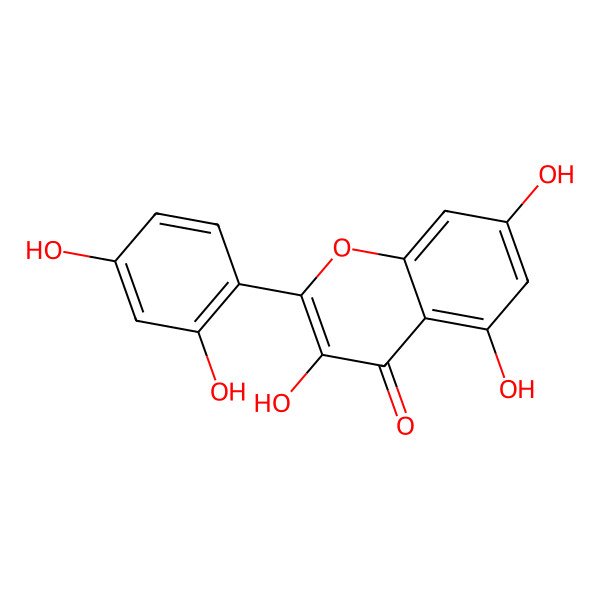
| IUPAC Name | 2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxychromen-4-one |
| SMILES (Canonical) | C1=CC(=C(C=C1O)O)C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O |
| SMILES (Isomeric) | C1=CC(=C(C=C1O)O)C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O |
| InChI | InChI=1S/C15H10O7/c16-6-1-2-8(9(18)3-6)15-14(21)13(20)12-10(19)4-7(17)5-11(12)22-15/h1-5,16-19,21H |
| InChI Key | YXOLAZRVSSWPPT-UHFFFAOYSA-N |
| Popularity | 5,907 references in papers |
| Molecular Formula | C15H10O7 |
| Molecular Weight | 302.23 g/mol |
| Exact Mass | 302.04265265 g/mol |
| Topological Polar Surface Area (TPSA) | 127.00 Ų |
| XlogP | 1.50 |
| 480-16-0 |
| Aurantica |
| Calico Yellow |
| 2',3,4',5,7-Pentahydroxyflavone |
| Al-Morin |
| Toxylon Pomiferum |
| 2-(2,4-Dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one |
| Morin hydrate |
| Bois d,Arc |
| Bois d'arc |
| There are more than 10 synonyms. If you wish to see them all click here. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| No predicted properties yet! | |||
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| CHEMBL1293255 | P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD+] |
31622.8 nM 35481.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL256 | P0DMS8 | Adenosine A3 receptor |
34000 nM |
Ki |
PMID: 8691424
|
| CHEMBL3577 | P00352 | Aldehyde dehydrogenase 1A1 |
19952.6 nM |
Potency |
via CMAUP
|
| CHEMBL2903 | P16050 | Arachidonate 15-lipoxygenase |
15848.9 nM |
Potency |
via CMAUP
|
| CHEMBL5393 | Q9UNQ0 | ATP-binding cassette sub-family G member 2 |
49000 nM 21000 nM |
IC50 IC50 |
PMID: 21354800
PMID: 21354800 |
| CHEMBL1293236 | P46063 | ATP-dependent DNA helicase Q1 |
28183.8 nM 39810.7 nM 15848.9 nM 7079.5 nM |
Potency Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP via CMAUP |
| CHEMBL2487 | P05067 | Beta amyloid A4 protein |
30300 nM |
IC50 |
PMID: 26719209
|
| CHEMBL4822 | P56817 | Beta-secretase 1 |
21700 nM 12300 nM |
IC50 IC50 |
DOI: 10.1007/s00044-012-0353-y
PMID: 21511472 |
| CHEMBL1293237 | P54132 | Bloom syndrome protein |
19952.6 nM 19952.6 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL4801 | P29466 | Caspase-1 |
39810.7 nM |
Potency |
via CMAUP
|
| CHEMBL3468 | P55210 | Caspase-7 |
15848.9 nM |
Potency |
via CMAUP
|
| CHEMBL4096 | P04637 | Cellular tumor antigen p53 |
3981.1 nM |
Potency |
via CMAUP
|
| CHEMBL3397 | P11712 | Cytochrome P450 2C9 |
10000 nM |
Potency |
via CMAUP
|
| CHEMBL340 | P08684 | Cytochrome P450 3A4 |
31622.8 nM 31622.8 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL2558 | P53355 | Death-associated protein kinase 1 |
11000 nM 1600 nM |
IC50 IC50 |
PMID: 26322379
PMID: 26322379 |
| CHEMBL2392 | P06746 | DNA polymerase beta |
562.3 nM 562.3 nM 4466.8 nM |
Potency Potency Potency |
via CMAUP
via Super-PRED via CMAUP |
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase |
5011.9 nM |
Potency |
via CMAUP
|
| CHEMBL4159 | Q99714 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein |
12589.3 nM 12589.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL242 | Q92731 | Estrogen receptor beta |
2927.18 nM |
IC50 |
via CMAUP
|
| CHEMBL1293267 | Q9HC97 | G-protein coupled receptor 35 |
1600 nM |
IC50 |
PMID: 24900447
|
| CHEMBL1293299 | Q03164 | Histone-lysine N-methyltransferase MLL |
35481.3 nM |
Potency |
via CMAUP
|
| CHEMBL4361 | Q07820 | Induced myeloid leukemia cell differentiation protein Mcl-1 |
2988.26 nM |
IC50 |
via CMAUP
|
| CHEMBL1293226 | B2RXH2 | Lysine-specific demethylase 4D-like |
28183.8 nM 35481.3 nM 25118.9 nM 39810.7 nM |
Potency Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP via CMAUP |
| CHEMBL2608 | P10253 | Lysosomal alpha-glucosidase |
3162.3 nM |
Potency |
via CMAUP
|
| CHEMBL1293224 | P10636 | Microtubule-associated protein tau |
112.2 nM 12589.3 nM 11220.2 nM 15848.9 nM 14125.4 nM 15848.9 nM 15848.9 nM 12589.3 nM |
Potency Potency Potency Potency Potency Potency Potency Potency |
via Super-PRED
via CMAUP via CMAUP via CMAUP via CMAUP via CMAUP via CMAUP via CMAUP |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit |
10000 nM 10000 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL2396508 | Q13332 | Receptor-type tyrosine-protein phosphatase S |
4300 nM |
IC50 |
PMID: 26602279
|
| CHEMBL3024 | P53350 | Serine/threonine-protein kinase PLK1 |
12600 nM |
IC50 |
PMID: 18762425
|
| CHEMBL6120 | Q96S37 | Solute carrier family 22 member 12 |
2000 nM |
IC50 |
PMID: 17325024
|
| CHEMBL1293256 | P40225 | Thrombopoietin |
3162.3 nM 3162.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL2599 | P43405 | Tyrosine-protein kinase SYK |
28000 nM |
IC50 |
PMID: 22257213
|
| CHEMBL1075138 | Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 |
199.5 nM 199.5 nM |
Potency Potency |
via Super-PRED
via CMAUP |
| CHEMBL1293227 | O75604 | Ubiquitin carboxyl-terminal hydrolase 2 |
18492.7 nM |
Potency |
via CMAUP
|
| CHEMBL1929 | P47989 | Xanthine dehydrogenase |
10100 nM |
IC50 |
PMID: 9461655
|
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 98.71% | 91.11% |
| CHEMBL1951 | P21397 | Monoamine oxidase A | 98.13% | 91.49% |
| CHEMBL3194 | P02766 | Transthyretin | 95.39% | 90.71% |
| CHEMBL1806 | P11388 | DNA topoisomerase II alpha | 94.72% | 89.00% |
| CHEMBL2581 | P07339 | Cathepsin D | 94.71% | 98.95% |
| CHEMBL1929 | P47989 | Xanthine dehydrogenase | 91.97% | 96.12% |
| CHEMBL2345 | P51812 | Ribosomal protein S6 kinase alpha 3 | 91.94% | 95.64% |
| CHEMBL242 | Q92731 | Estrogen receptor beta | 91.83% | 98.35% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 90.32% | 95.56% |
| CHEMBL1860 | P10827 | Thyroid hormone receptor alpha | 85.90% | 99.15% |
| CHEMBL2635 | P51452 | Dual specificity protein phosphatase 3 | 85.69% | 94.00% |
| CHEMBL3401 | O75469 | Pregnane X receptor | 85.51% | 94.73% |
| CHEMBL5845 | P23415 | Glycine receptor subunit alpha-1 | 85.04% | 90.71% |
| CHEMBL1293249 | Q13887 | Kruppel-like factor 5 | 84.96% | 86.33% |
| CHEMBL2073 | P07947 | Tyrosine-protein kinase YES | 83.55% | 83.14% |
| CHEMBL4208 | P20618 | Proteasome component C5 | 82.82% | 90.00% |
| CHEMBL3864 | Q06124 | Protein-tyrosine phosphatase 2C | 81.70% | 94.42% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.
| PubChem | 5281670 |
| NPASS | NPC216361 |
| ChEMBL | CHEMBL28626 |
| LOTUS | LTS0113386 |
| wikiData | Q418224 |