| 63-68-3 |
| methionine |
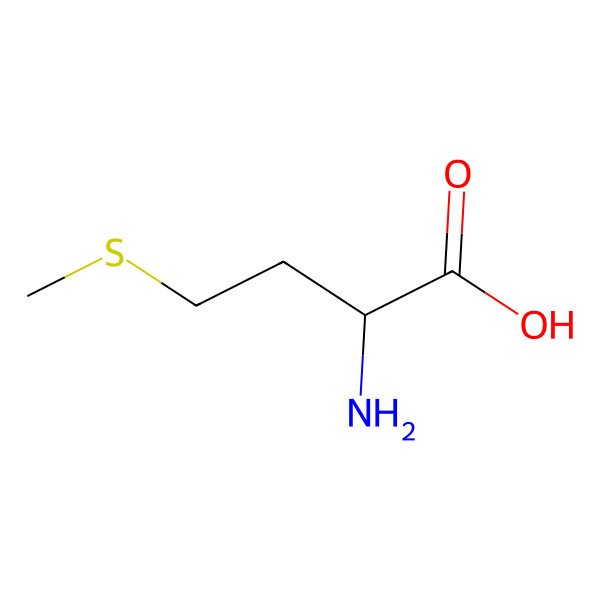
| h-Met-oh |
| (S)-2-Amino-4-(methylthio)butanoic acid |
| Cymethion |
| Liquimeth |
| L-(-)-Methionine |
| S-Methionine |
| Neo-methidin |
| L-Methioninum |
| Methilanin |
| (L)-Methionine |
| Acimethin |
| (S)-methionine |
| metionina |
| Methionine (VAN) |
| Methioninum |
| (2S)-2-amino-4-(methylsulfanyl)butanoic acid |
| Metionina [DCIT] |
| L-Methionin |
| L-alpha-Amino-gamma-methylmercaptobutyric acid |
| h-Met-h |
| Methioninum [INN-Latin] |
| L-Homocysteine, S-methyl- |
| met |
| CCRIS 5528 |
| CCRIS 5536 |
| HSDB 4317 |
| L(-)-Amino-gamma-methylthiobutyric acid |
| L-Met |
| 2-Amino-4-methylthiobutanoic acid (S)- |
| L-alpha-Amino-gamma-methylthiobutyric acid |
| L-gamma-Methylthio-alpha-aminobutyric acid |
| 2-Amino-4-(methylthio)butyric acid, (S)- |
| METHIONINE, L- |
| Butanoic acid, 2-amino-4-(methylthio)-, (S)- |
| Methionine [USAN:INN] |
| UNII-AE28F7PNPL |
| (S)-2-amino-4-(methylthio)butyric acid |
| AE28F7PNPL |
| Toxin WAR (Bacillus thuringiensis strain PS205C) |
| EINECS 200-562-9 |
| (2S)-2-amino-4-methylsulfanyl-butanoic acid |
| C-11 Methionine |
| NSC 22946 |
| gamma-Methylthio-alpha-aminobutyric acid |
| CHEBI:16643 |
| L-a-Amino-g-methylthiobutyric acid |
| carbon-11 methionine |
| S-Methyl-L-homocysteine |
| (S)-(+)-Methionine |
| MFCD00063097 |
| NSC-22946 |
| (2S)-2-Amino-4-methylsulfanylbutanoic acid |
| C-11 Met |
| CHEMBL42336 |
| DTXSID5040548 |
| L-2-Amino-4methylthiobutyric acid |
| Methionine [USAN:USP:INN:BAN] |
| meritonin |
| (35S)Methionine |
| L-Methionine-34S |
| METHIONINE (II) |
| METHIONINE [II] |
| METHIONINE (MART.) |
| METHIONINE [MART.] |
| Hmet |
| 1006386-95-3 |
| METHIONINE (EP MONOGRAPH) |
| METHIONINE [EP MONOGRAPH] |
| METHIONINE (USP MONOGRAPH) |
| METHIONINE [USP MONOGRAPH] |
| LEUCINE IMPURITY B (EP IMPURITY) |
| LEUCINE IMPURITY B [EP IMPURITY] |
| 58576-49-1 |
| 2-amino-4-(methylthio)-butyric acid |
| l-2-Amino-4-(methylthio)butyric acid |
| (2S)-2-amino-4-(methylsulfanyl)butanoate |
| L-2-Amino-4-(methylthio)butanoic acid |
| L-metionina |
| (C5-H11-N-O2-S)x- |
| L-Lobamine |
| 3h-l-methionine |
| g-Methylthio-a-aminobutyric acid |
| racemic methionine |
| 1wkm |
| NSC 118113 |
| D-2-Amino-4-(methylthio)butanoic acid |
| Toxin WAR |
| Butyric acid, 2-amino-4-(methylthio)- |
| 2-Amino-4-(methylthio)butyrate |
| Methionine (USP) |
| L(-)-Methionin |
| a-Amino-g-methylmercaptobutyric acid |
| methioninum racemicum |
| L-Methionine,(S) |
| (R)-2-Amino-4-(methylmercapto)butyric acid |
| 1pg2 |
| 1qq9 |
| L-Methionine Z (TN) |
| METHIONINE [MI] |
| L-Methionine (JP17) |
| METHIONINE [INN] |
| 128488-79-9 |
| METHIONINE [HSDB] |
| METHIONINE [INCI] |
| METHIONINE [USAN] |
| (S)-2-Amino-4-(methylmercapto)butyric acid |
| Methionine (L-Methionine) |
| METHIONINE [VANDF] |
| Methionine, L- (8CI) |
| bmse000044 |
| bmse000915 |
| D0PO8E |
| L-METHIONINE [FCC] |
| L-METHIONINE [JAN] |
| SCHEMBL4226 |
| G-methylthio-a-aminobutyrate |
| L-Methionine (H-Met-OH) |
| METHIONINE [WHO-DD] |
| 2-Amino-4-methylthiobutanoate |
| L-a-amino-g-methylthiobutyrate |
| L-METHIONINE [USP-RS] |
| GTPL4814 |
| A-amino-g-methylmercaptobutyrate |
| IS_METHIONINE-METHYL-D3 |
| DTXCID3020548 |
| SCHEMBL15702352 |
| V03AB26 |
| Pharmakon1600-01301006 |
| alpha-amino-alpha-aminobutyric acid |
| gamma-methylthio-alpha-aminobutyrate |
| HY-N0326 |
| L-2-Amino-4-methylthiobutyric acid |
| BDBM50142500 |
| MFCD00801344 |
| NSC760117 |
| s5633 |
| L-alpha-amino-gamma-methylthiobutyrate |
| L-Methionine, Vetec(TM), 98.5% |
| (S)-2-Amino-4-(methylthio)butanoate |
| AKOS000281626 |
| AKOS015852512 |
| L-Methionine, labeled with carbon-11 |
| alpha-amino-gamma-methylmercaptobutyrate |
| CCG-266196 |
| CS-W020566 |
| DB00134 |
| LS-2338 |
| NSC-760117 |
| (S)-2-amino-4-(methylthio)-Butanoate |
| NCGC00160620-01 |
| NCGC00160620-02 |
| AS-10898 |
| BP-31235 |
| (S)-2-amino-4-(methylthio)-Butanoic acid |
| L-Methionine, BioUltra, >=99.5% (NT) |
| A5456 |
| AM20100552 |
| M0099 |
| EN300-52631 |
| L-Methionine, SAJ special grade, >=98.5% |
| C00073 |
| D00019 |
| D70895 |
| L(-)-amino-alpha-amino-alpha-aminobutyric acid |
| L-Methionine, reagent grade, >=98% (HPLC) |
| M-3100 |
| M02939 |
| M02945 |
| L-Methionine, Vetec(TM) reagent grade, >=98% |
| A934626 |
| L-Methionine, Cell Culture Reagent (H-L-Met-OH) |
| C6CB5837-2B49-4B25-AAB0-D305DAFE26EB |
| Q22124685 |
| F1905-8241 |
| Z756440050 |
| L-Methionine, certified reference material, TraceCERT(R) |
| Methionine, European Pharmacopoeia (EP) Reference Standard |
| N-(2CT Resin)-L-Met-OH (200-400 mesh, > 0.3 mmol/g) |
| L-Methionine, United States Pharmacopeia (USP) Reference Standard |
| L-Methionine, Pharmaceutical Secondary Standard; Certified Reference Material |
| Soft tissue sarcoma-associated protein (human clone WO2004048938-SEQID-1139) |
| L-Methionine, from non-animal source, meets EP, JP, USP testing specifications, suitable for cell culture, 99.0-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|